Geoduck Larval DNA Extractions
Extracted DNA from Larval Geoduck Tissue frozen at -80°C and stored in RNALater
Sample list 20170106
- EPI-1
- EPI-2
- EPI-3
- EPI-4
- EPI-5
- EPI-6
- EPI-7
- EPI-8
- EPI-9
- EPI-11
- EPI-12
- EPI-13
- EPI-14
- EPI-15
- EPI-16
- EPI-17
- EPI-18
- EPI-19
- EPI-20
- EPI-21
- EPI-22
- EPI-23
- EPI-24
- EPI-25
- EPI-26
- EPI-27
- EPI-28
- EPI-29
- EPI-30
- EPI-31
- EPI-32
- EPI-33
- EPI-34
- EPI-35
- EPI-36
- EPI-37
- EPI-39
- EPI-40
- EPI-60
-
EPI-66
- Had a lot of difficulty concentrating #1. It seems this sample may have been stored in ethanol and not RNA later
DNA Extractions
- added 180µl of Buffer ATL and 20µl of Proteinase K
- heated samples at 56°C for 1hr
- added 200µl Buffer AT and heated an additional 10min
- added 200µl of 100% ethanol and proceeded to column stage
- Proceeded with DNA Extraction Protocol
- Did not do the RNase treatment
- Eluted in 125µl of AE Buffer
DNA Quantification
- Used 1µl of sample and 199µl of Qubit Mix
- Ran Qubit dsDNA BR DNA Quantification Protocol
DNA Concentrations
| Sample.ID | DNA Conc(ng/µl) |
|---|---|
| EPI_1 | 0 |
| EPI_2 | 4.16 |
| EPI_3 | 57.8 |
| EPI_4 | 91.2 |
| EPI_5 | 0 |
| EPI_6 | 0 |
| EPI_7 | 0 |
| EPI_8 | 56.4 |
| EPI_9 | 0 |
| EPI_11 | 3.06 |
| EPI_12 | 41.2 |
| EPI_13 | 2.98 |
| EPI_14 | 53.4 |
| EPI_15 | 71.6 |
| EPI_16 | 62.4 |
| EPI_17 | 8.42 |
| EPI_18 | 51.8 |
| EPI_19 | 46.2 |
| EPI_20 | 68.4 |
| EPI_21 | 36.2 |
| EPI_22 | 98 |
| EPI_23 | 8.64 |
| EPI_24 | 104 |
| EPI_25 | 8.22 |
| EPI_26 | 71.2 |
| EPI_27 | 36.6 |
| EPI_28 | 71 |
| EPI_29 | 21 |
| EPI_30 | 79.6 |
| EPI_31 | 10.8 |
| EPI_32 | 80.8 |
| EPI_33 | 39.4 |
| EPI_34 | 96.6 |
| EPI_35 | 18.1 |
| EPI_36 | 80 |
| EPI_37 | 15.3 |
| EPI_39 | 22.8 |
| EPI_40 | 67 |
| EPI_60 | 161 |
| EPI_62 | 186 |
- estimated total DNA from a sample of 115µl after Qubit and gel
DNA Quality
Ran a quality check of DNA using a 1.5% TAE gel in 10x TAE running buffer
Gel Preparation
1X TAE
- 40 mM Tris (pH 7.6)
- 20 mM acetic acid
-
1 mM EDTA
- Added 1.5g of Agarose to 150ml of 10X TAE and heated until clear
- Added 12µl of Ethidium Bromide to gel
- Poured gel with 16 upper wells and 16 lower wells
- Once gel was set, Added 6µl of 6x loading dye to each sample of 6µl of DNA (6x purple loading dye #B70245 New Endland BioLabs) and added 8µl of mix to each well
- Ran gel at 100v for 60 minutes
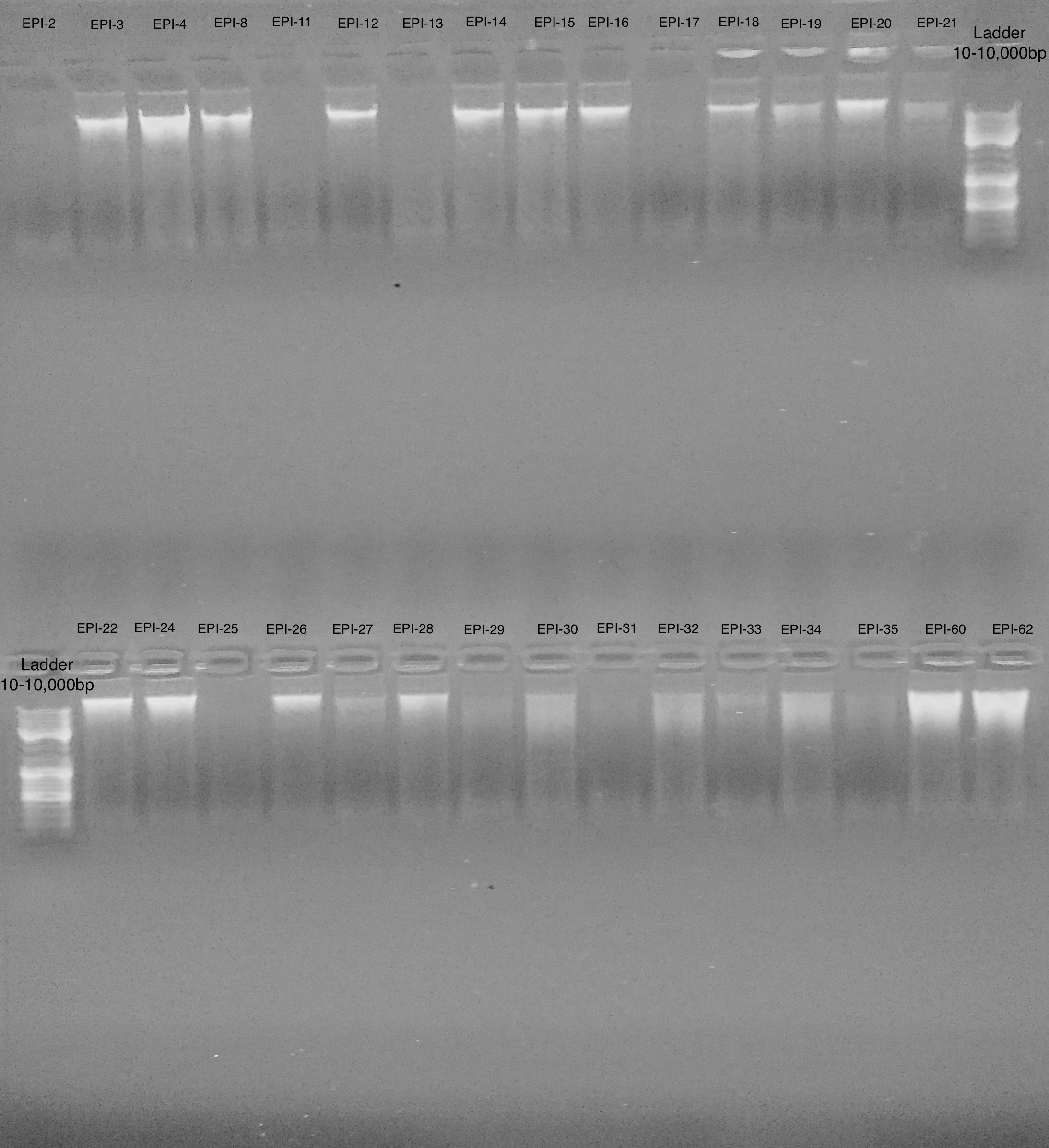
Samples
Gel Top
- EPI-2
- EPI-3
- EPI-4
- EPI-8
- EPI-11
- EPI-12
- EPI-13
- EPI-14
- EPI-15
- EPI-16
- EPI-17
- EPI-18
- EPI-19
- EPI-20
- EPI-21
- O Gene Ruler Mix SM 1173 Thermo Fisher 0.1µg/µl Range 100–10,000 bp
Gel Bottom
- O Gene Ruler Mix SM 1173 Thermo Fisher 0.1µg/µl Range 100–10,000 bp
- EPI-22
- EPI-23
- EPI-24
- EPI-25
- EPI-26
- EPI-27
- EPI-28
- EPI-29
- EPI-30
- EPI-31
- EPI-32
- EPI-33
- EPI-34
- EPI-35
- EPI-60
- EPI-62
Gel Setup
Gel 1
Conclusions
-
Most samples have DNA with large fragment size and minimal degradation. Samples with ~<10ng/µl are not detectable on gel.
-
Samples will be processed for RADseq and EPIRADseq according to previous prep by Jay Dimond JD RAD Days1-2 JD RAD Days3-4 JD RAD Day5
Sample Selection
Samples were selected from Sample List
Day 0 Ambient pCO2
- EPI-3
- EPI-4
Day 6 Ambient pCO2
- EPI-33
- EPI-34
- EPI-35
Day 6 High pCO2
- EPI-29
- EPI-30
- EPI-32
Day 10 Ambient pCO2
- EPI-60
Day 10 High pCO2
Written on January 6, 2017
