Geoduck Juvenile and Larval DNA Extractions (part5)
Extracted DNA from Juvenile Geoduck Tissue frozen at -80°C Extracted DNA from Larval Geoduck Tissue frozen at -80°C and stored in RNALater
Whole issue was previously homogenized using the Sample Homogenization Protocol
Sample list 20161027
- EPI-42
- EPI-45
- EPI-46
- EPI-47
- EPI-48
- EPI-50
- EPI-103
- EPI-143
- EPI-161
- EPI-242
- EPI-243
- EPI-247
- EPI-258
- EPI-271
- EPI-272
- EPI-283
- EPI-298
- EPI-1
- EPI-10
- EPI-36
- EPI-38
- EPI-65
- EPI-66
-
EPI-78
- Had a lot of difficulty concentrating #1 and #78 to remove liquid before extraction. Sample 78 consumed, sample 1 available at -20.
DNA Extractions
- added ~40mg of tissue to 180µl of Buffer ATL and 20µl of Proteinase K
- for samples 65 and 66 there was so much material used 4x the buffer and protK
- took 1 180µl aliquot forward into extraction and saved rest of EPI-65 and EPI-66 in ATL buffer at room temp
-
Also saved EPI-36 and EPI-38 pellets in new 180µl of ATL buffer at room temp
- Proceeded with DNA Extraction Protocol
- Did not do the RNase treatment
- Eluted in 150µl of AE Buffer
- Saved samples at -80°C in 2 aliquots (6µl for gel and Qubit,~140µl)
DNA Quantification 20161028
- Used 2µl of sample and 198µl of Qubit Mix
- Ran Qubit dsDNA BR DNA Quantification Protocol
DNA Concentrations
| Sample.ID | Qubit Conc(ng/µl) | Dilution | Initial Conc(ng/µl) |
|---|---|---|---|
| EPI-42 | 79 | 1 | 79 |
| EPI-45 | 43.1 | 1 | 43.1 |
| EPI-46 | 21.2 | 1 | 21.2 |
| EPI-47 | 57.6 | 1 | 57.6 |
| EPI-48 | 12.6 | 1 | 12.6 |
| EPI-50 | 8.21 | 1 | 8.21 |
| EPI-103 | 18.3 | 1 | 18.3 |
| EPI-143 | 41.2 | 1 | 41.2 |
| EPI-161 | 24.9 | 1 | 24.9 |
| EPI-242 | 15.5 | 1 | 15.5 |
| EPI-243 | 17.2 | 1 | 17.2 |
| EPI-247 | 8.23 | 1 | 8.23 |
| EPI-258 | 18.8 | 1 | 18.8 |
| EPI-271 | 39.1 | 1 | 39.1 |
| EPI-272 | 105 | 1 | 105 |
| EPI-283 | 36.6 | 1 | 36.6 |
| EPI-298 | 79.9 | 1 | 79.9 |
| EPI-1 | 0 | 1 | 0 |
| EPI-10 | 42.6 | 1 | 42.6 |
| EPI-36 | 80 | 1 | 80 |
| EPI-38 | 74.2 | 1 | 74.2 |
| EPI-65 | 3 | 1 | 3 |
| EPI-66 | 11.2 | 1 | 11.2 |
| EPI-78 | 3.13 | 1 | 3.13 |
- a 140µl aliquot with an average of ~35ng/µl has ~4.9µg of DNA
DNA Quality 20161028
Ran a quality check of DNA using a 1% TAE gel in 10x TAE running buffer
** Accidentally used 10x TAE, which resulted in gel running very slowly
Gel Preparation
1X TAE
- 40 mM Tris (pH 7.6)
- 20 mM acetic acid
- 1 mM EDTA
Accidentally used 10x TAE, which resulted in gel running very slowly
Samples
Gel Top
- O Gene Ruler Mix SM 1173 Thermo Fisher 0.1µg/µl Range 100–10,000 bp
- EPI-42
- EPI-45
- EPI-46
- EPI-47
- EPI-48
- EPI-50
- EPI-103
- EPI-143
- EPI-161
- EPI-242
- EPI-243
- EPI-247
- EPI-258
- EPI-271
- O Gene Ruler Mix SM 1173 Thermo Fisher 0.1µg/µl Range 100–10,000 bp
Gel Bottom
- O Gene Ruler Mix SM 1173 Thermo Fisher 0.1µg/µl Range 100–10,000 bp
- EPI-272
- EPI-283
- EPI-298
- EPI-1
- EPI-10
- EPI-36
- EPI-38
- EPI-65
- EPI-66
- EPI-78
- O Gene Ruler Mix SM 1173 Thermo Fisher 0.1µg/µl Range 100–10,000 bp
Gel Setup
- Added 1.5g of Agarose to 150ml of 10X TAE and heated until clear
- Added 12µl of Ethidium Bromide to gel
- Poured gel with 16 upper wells and 16 lower wells
- Once gel was set, Added 4µl of 6x loading dye to each sample of 4µl of DNA (6x purple loading dye #B70245 New Endland BioLabs) and added 8µl of mix to each well
- Ran gel at 100v for 60 minutes
Gel 1
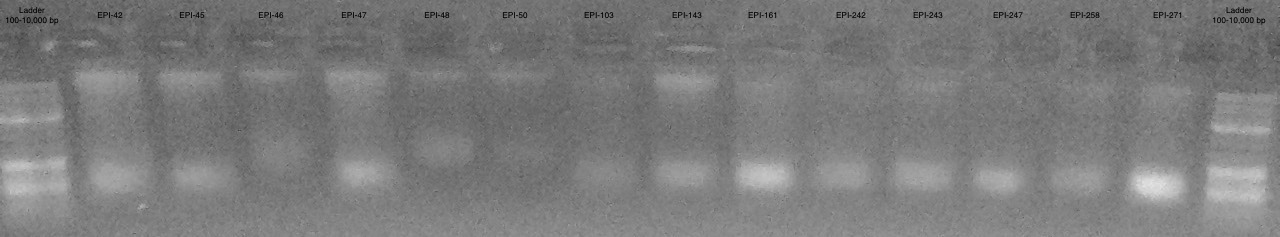
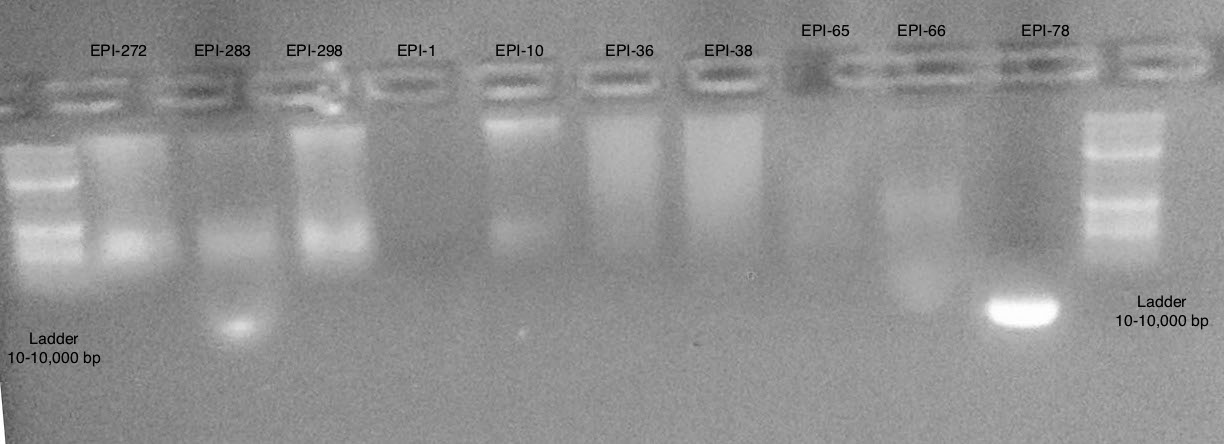
Conclusions
- Samples are more difficult to see then on actual gel
- Appears to be some large band >10,000 bp and some smear below
- Most samples have high molecular weight DNA present
- Some are too low of concentration to see (EPI-1, EPI-65)
- EPI-78 appears to only be very small degraded fragments
-
EPI-36 and EPI-38 appear to have degradation in comparison to EPI-10
- Will continue to move forward with DNeasy DNA Extraction Protocol
Written on October 28, 2016
