Geoduck RRBS Testing
Testing RRBS protocol using MSPI cutting, BS treatment, and Illumina Trueseq DNA methylation kit
MSPI enzyme cuts between Cs at CCGG sites in the genome. When bisulfite treatment is used after cutting, data can be gathered from the beginning of the read CG and at other CGs within the sequence read lenght (~100bp). This cutting and conversion creates the imput material for the library prep. The concern is that the Illumina Trueseq DNA methylation kit was optimized for whole genome library prep and the DNA following the MSPI cutting and BS treatmetn might have the wrong size and quantitify of frgaments to generate good seqeuncing results.
I am testing this process with 3 samples with 1µg of DNA from each.
Step 1 MSPI cutting
Mix the following and incubate overnight at 37°C.
| Reagents | Epi 272 | Epi 298 | Epi 42 |
|---|---|---|---|
| NEBuffer2 10x | 3µl | 3µl | 3µl |
| MSPI 20U/µl | 1µl | 1µl | 1µl |
| H20 | 16.5µl | 13.5µl | 13.3µl |
| 1µg DNA | 9.5µl | 12.5µl | 12.7µl |
| total | 30µl | 30µl | 30µl |
Step 2 BS treatment
EZ DNA Methylation-Gold Kit (Catalog Nos. D5005 & D5006) EZ DNA Methylation-Gold Manual
| Input Material | 11/30 Sample.ID | Volume | Spike |
|---|---|---|---|
| EPI-42 | 42 L | 20µl of MSPI cut DNA | Lambda DNA Added (0.1%) |
| EPI-272 | 272 L | 20µl of MSPI cut DNA | Lambda DNA Added (0.1%) |
| EPI-298 | 298 L | 20µl of MSPI cut DNA | Lambda DNA Added (0.1%) |
| EPI-42 | 42 N | 10µl of MSPI cut DNA + 10µl of water | No |
| EPI-272 | 272 N | 10µl of MSPI cut DNA + 10µl of water | No |
| EPI-298 | 298 N | 10µl of MSPI cut DNA + 10µl of water | No |
| EPI-272 | 272 WG | 6.27µl genomic DNA + 13.73µl H20 | Lambda DNA Added (~0.1%) |
- Added 20µl of sample described in table to 130 µl of CT Conversion Reagent, mixed by flicking, and spun down
- Placed tubes in PCR machine set as follows:
- 98°C for 10 min
- 64°C for 2.5 hours
- 4°C forever
- Added 600µl of M-Binding Buffer and 150µl of sample to IC Column, mixed by inversion, and spun at 15,000g for 30 sec, and discarded flow through
- Added 100µl of M-Wash Buffer spun at 15,000g for 30 sec, and discarded flow through
- added 200µl of **M-Desulphonation Buffer to column and incubate at room temp for 15min and spun at 15,000g for 30 sec, and discarded flow through
- Added 200µl of M-Wash Buffer spun at 15,000g for 30 sec, and discarded flow through
- Added 200µl of M-Wash Buffer spun at 15,000g for 30 sec, and discarded flow through
- Placed column in 1.5ml centrifuge tube and added 10µl of M-Elution Buffer to the column spun at 15,000g for 30 sec, repeated for a second elution of 10µl
Check of DNA size and suantity
Quality
Ran 1µl of each sample on the RNA Pico chip to quant ssDNA following bisulfite conversion BS converson results
The comparision of the same sample is highlighted in yellow. MSPI cutting and lambda DNA addition (top), MSPI cutting and no lambda DNA addition (middle), Genomic DNA, no cutting, and lambda DNA addition (bottom).
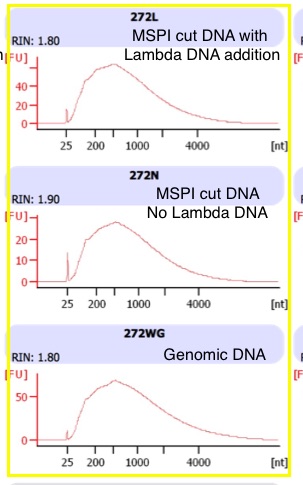
The peaks in my samples are ~700bp, whereas the image from the illumina prep guide shows a a peak at ~1000
Quantity
| Sample.ID | ng.µl |
|---|---|
| 42L | 38.78 |
| 272L | 38.16 |
| 298L | 43.89 |
| 42N | 24.35 |
| 272N | 22.44 |
| 298N | 25.16 |
| 272WG | 38.58 |
- There seems to be no effect of MSPI versus whole genome on the size of the DNA fragments after BS conversion
- There is no effect of the MSPI cutting on total quantity
- The major concern is that in all the samples the peak size is smaller than the example in the illumina kit. It is possible there was initial degradation or degradation during BS conversion that would shift the peak to the left (smaller). I talked to Illumina and they recommeded checking initial DNA quality again and starting with 100ng instead of 1000ng of input material.
