Poc ID via PocHistone RFLP
Pocillopora species ID PocHistone PCR and RFLP
10 May 2024
Prepare Reagents
Primers - Johnston et al. 2018
- Forward primer (PocHistoneF: 5′-ATTCAGTCTCACTCACTCACTCAC- 3′)
- Reverse primer (PocHistoneR: 5′-TATCTTCGAACAGACCCACCAAAT-3′)
- Master Mix EmeraldAmp GT PCR Master Mix
Primers were resuspended to 100µM and diluted to 10µM for use in PCR.
Sample List
Recruit TPC
REC-001 REC-002 REC-003 REC-004 REC-005 REC-008 REC-009 REC-010 REC-011 REC-012 REC-013 REC-014 REC-015 REC-016 REC-017 REC-018 REC-019 REC-020 REC-021 REC-022 REC-023 REC-024 REC-025 REC-027 REC-028 REC-029 REC-030 REC-031 REC-032 REC-033 REC-034 REC-035 REC-037 REC-038 REC-039 REC-040
Histology Samples
POC-466 POC-467 POC-468 POC-469 POC-475 POC-476 POC-477
Flow Samples
FLOW-007 FLOW-008 FLOW-009 FLOW-010 FLOW-011 FLOW-019 FLOW-020 FLOW-021 FLOW-022 FLOW-023 FLOW-025 FLOW-030 FLOW-031 FLOW-032 FLOW-033 FLOW-034 FLOW-026 FLOW-027 FLOW-028 FLOW-029
PocHistone PCR
EmeraldAmp GT PCR Master Mix (2x).
Master Mix: For 65 rxns + 3 = 68
| Reagent | 1Rxn µl | 68 Rxn µl |
|---|---|---|
| EmeraldAmp GT Mix (2x) | 12.5 | 850 |
| F primer PocHistoneF (10µM) (10µM) | 0.3 | 20.4 |
| R primer PocHistoneR (10µM)(10µM) | 0.3 | 20.4 |
| H2O | 10.9 | 741.2 |
| DNA | 1 | NA |
| Total volume | 25 | 450 |
This was not enough for the last 2 samples, so those were each mixed with 1x volumes individually. Flow-0028 Flow-0029
Thermal Cycling Conditions
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 60 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
PocHistone PCR Gel and RFLP
Ran PocHIstone RFLP with NEB Restriction Enzyme XhoI, NEB Cat# R0146S
- Restriction Enzyme XhoI R0146Sk
- Restriction Enzyme Buffer rCutSmart B6004S
Restriction Digest Master Mix
Reagent |1Rxn µl |80 Rxn µl
—|—|—
XhoI restriction enzyme | 0.5 |40
1X CutSmartR buffer |0.5| 40
For each sample added 1µl of Restriction Digest Master Mix to 15µl of PCR product. Incubated samples for 1 hr at 37°C followed by 20 min at 65°C to inactivate the enzymes.
PCR Gel protocol
Prepared two medium gels using 100ml of 1xTAE + 1.5g Agarose, melted gel in the microwave and let cool and added 1.5µl Gelgreen and let harden. Ran gel at 60V for 110 minutes in 1x TAE.
Gel Samples
GeneRuler 100 bp DNA Ladder Thermo FIsher Cat SM024
Positive control = P. grandis 198
Gel Results
Bands of 669bp indicates the samples are P. meandrina. Two bands at 287 and 382 indicate the species is P. grandis (P. eydouxi).
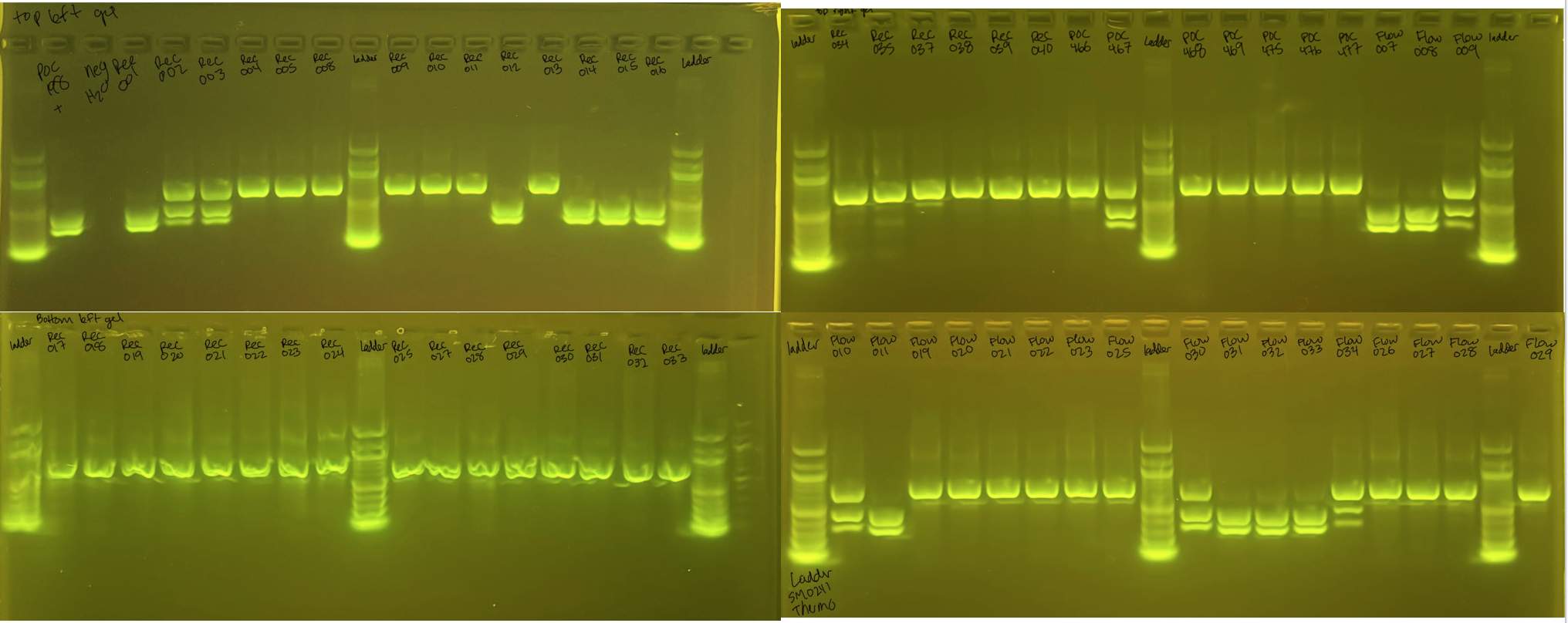
POC-476 = P. tua from Sanger Seq (Haplotype 10). Does not cut with XhoI.
POC-477 = P. tua from Sanger Seq (Haplotype 10). Does not cut with XhoI.
P. meandrina 1 band
REC-004 REC-005 REC-008 REC-009 REC-010 REC-011 REC-013 REC-017 REC-018 REC-019 REC-020 REC-021 REC-022 REC-023 REC-024 REC-025 REC-027 REC-028 REC-029 REC-030 REC-031 REC-032 REC-033 REC-034 REC-035 REC-037 REC-038 REC-039 REC-040 POC-466 POC-468 POC-469 POC-475 FLOW-019 FLOW-020 FLOW-021 FLOW-022 FLOW-023 FLOW-024 FLOW-025 FLOW-026 FLOW-027 FLOW-028 FLOW-029
P. grandis 2 bands
POC-198 = positive control REC-001 REC-012 REC-014 REC-015 REC-016 FLOW-007 FLOW-008 FLOW-011 FLOW-031 FLOW-032 FLOW-033
Unknown 3 bands
REC-002 REC-003 POC-467 FLOW-009 FLOW-010 FLOW-030 FLOW-034
All of these 3 band FLOW samples are from the P. grandis morphology. I will Sanger sequence all of the 3 band samples next to confirm if they are Pgrandis or a different species. It is possible the 3 bands come from incomplete cutting of the PocHistone product by the XhoI, as the 3 bands are in very similar positions to the uncut and cut products.
