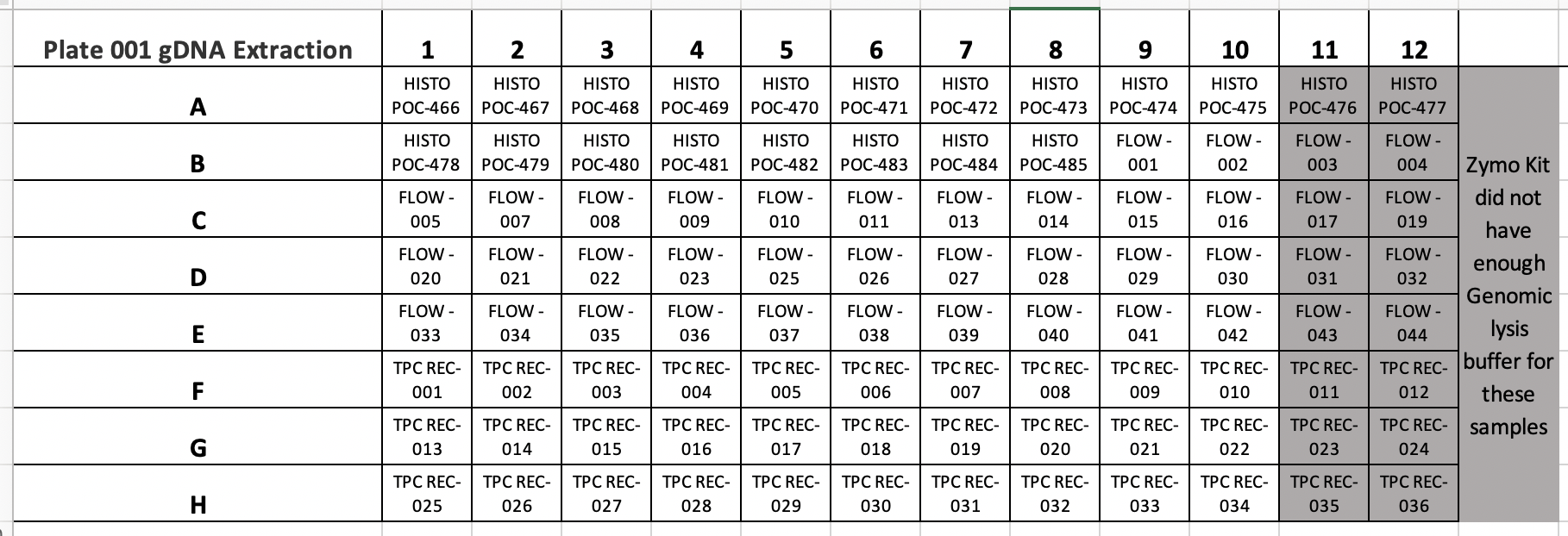
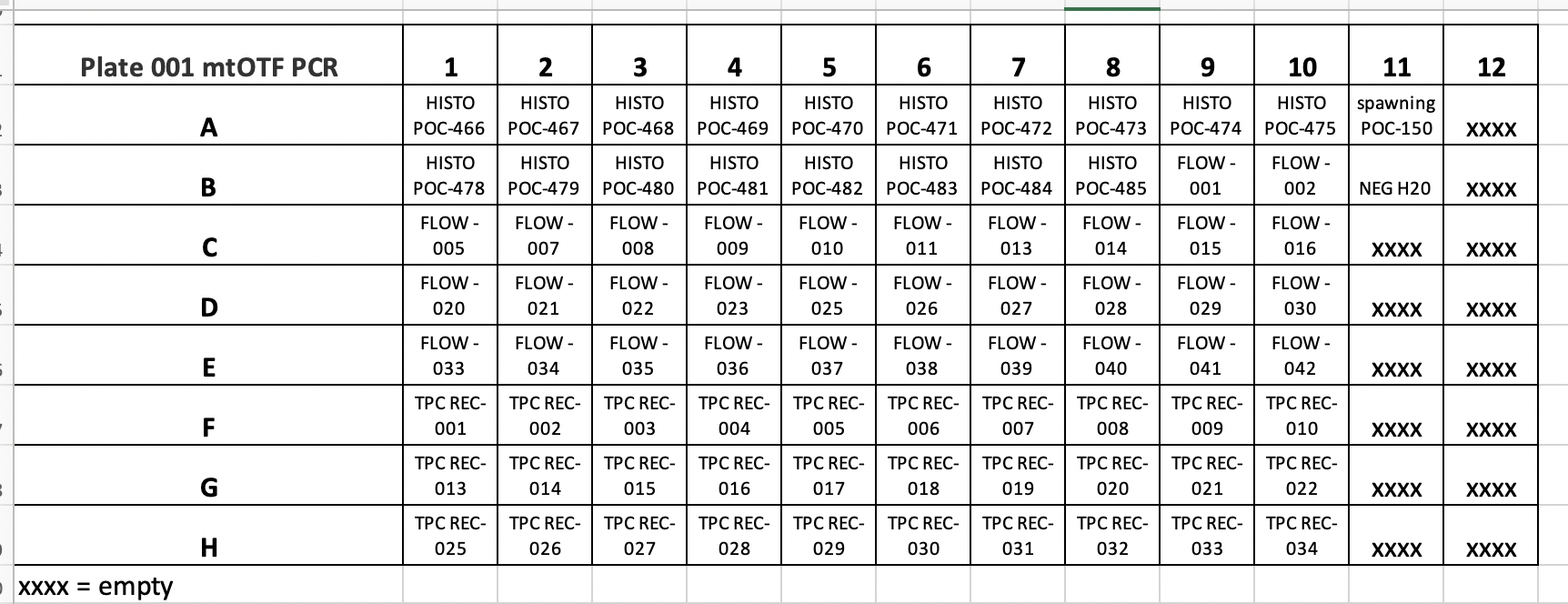
Poc ID via mtORF POC RAPID Plate 001
Pocillopora species ID COTS POC RAPID Plate 001
Equipment and Reagents
- Sample Preservation and Lysis Zymo Research DNA/RNA Shield R1100-250
- DNA Extraction 96 Well Plate Zymo Research Quick-DNA™ 96 D3010
- DNA Extraction Individual Samples Zymo Research Quick-DNA™ Miniprep plus kit extraction Cat D4068
- Proteinase K Zymo Proteinase K with Storage BufferZymo D3001-2-20
- PK Digestion Buffer Zymo PK Digestion Buffer R1200-1-20
- Foward primer FatP6.1 200µM Stock IDT
- Reverse primer RORF 200µM Stock IDT
- Master Mix EmeraldAmp GT PCR Master Mix RR320A
- EmeraldAmp GT PCR Master Mix is a loading-dye-added version of EmeraldAmp MAX PCR Master Mix that is optimized for great performance and convenience in both standard and high-throughput PCR applications.
- Loading Dye NEB 6X Purple Loading Dye NEB Cat # B7024S
- Gel Stain Biotium GelGreen Nucleic Acid Gel Stain, 10,000X in Water Fisher Cat NC9728313
- DNA Ladder Image NEB Cat N0550G 1kb Gel ladder NEB Cat N0550G
- DNA Ladder NEB Cat N0550G 1kb Gel ladder NEB Cat N0550G
- KapaPure Beads Fisher 50-196-5220 Roche Diagnostics 07983280001
- Eppendorf Deepwell Plate 96/2000 µL plates Eppendorf Cat # 951033502
11 April 2024
DNA Extraction
DNA was extracted withZymo Research Quick-DNA™ 96 Kit D3010
Used Page 10 Proteinase K Digestion with DNA/RNA Shield
Used Eppendorf Deepwell Plate 96/2000 µL plates Catalog No. 951033502 for reagent and sample mixing prior to adding to Zymo column plate
Need Proteinase K with Storage Buffer (Zymo D3001-2-20) and PK Digestion Buffer (Zymo R1200-1-20)
For each of 3 tubes of Proteinase K with Storage Buffer (Zymo D3001-2-20) I added 1040µl of Proteinas K Storage Buffer to lyophilized Proteinase K 20mg tube and mixed by vortex.
-
First 150µl of new DNA/RNA Shield was added to each well in the Eppendorf 96 Deepwell plate
-
Next 30µl of PK Digestion buffer and 15µl of resusdended Proteinase K solution was added to each well.
-
Next 150µl of sample added to each well and samples were mixed with solution.
The samples were extracted according to the manufacturer’s instructions for the Quick DNA 96 Kit for samples stored in DNA/RNA Shield including the addition of 15µl of Proteinase K (20mg ml-1).
Zymo Kit Genomic Lysis Buffer was prepared according to Instructions Page 4. 500µl of beta-mercaptoethanol was added to the 100ml bottle of Genomic Lysis Buffer.
- Next 4 volumes of Zymo Kit Genomic Lysis Buffer was added for each volume of the sample digestion (e.g., add 1200µl of Genomic Lysis Buffer to 300µl of sample digestion). Samples were mixed via pipetting and the mixutres were incubated for 5 minutes at room temperature.
MASSIVE PROBLEM HERE IN ZYMO INSTRUCTIONS FOR THE KIT INSTRUCTIONS PROVIDED. THERE IS ONLY 100ML OF GENOMIC LYSIS BUFFER PROVIDED, BUT THE KIT INSTRUCTIONS DO NOT TELL YOU TO ORDER MORE PRIOR TO STARTING THE KIT. THEREFORE WE WERE LEFT WITH NO GENOMIC LYSIS BUFFER FOR COLUMNS 11 AND 12 OF PLATE 001. THEREFORE WE MOVED FORWARD WITH EXTRACTIONS OF 80 SAMPLES FROM PLATE 001 COLUMNS 1-10 ALL ROWS. COLUMNS 11 AND 12 WERE MOVED TO NEW INDIVIDUALLY LABELLED TUBES AND STORED IN THE -20°C FREEZER.

Each well of the Silicon-A plate set on the collection plate can hold 600µl, but this brings the liquid too close to the top and increases chances of carry over to other wells. Becasue of this, I added the sample in 4 steps: 1) 400µl and then 5 minute spin at 1000 rcf 1) 400µl and then 5 minute spin at 1000 rcf 1) 400µl and then 5 minute spin at 2500 rcf 1) 400µl and then 5 minute spin at 2500 rcf
The waste was removed from the collection plate after each centrifugation step.
-
Next 200µl of DNA Pre-wash buffer was added to each well and then the plate was spun for 5 minute at 2500 rcf
-
Next 300µl of g-DNA wash buffer was added to each well and then the plate was spun for 5 minute at 2500 rcf
-
DNA was then eluted in 60µl of kit Elution Buffer warmed to 70°C. DNA extractions were completed on 11 April 2024 and DNA stored at -20°C in the elution plate covered with a foil seal.
Samples
12 April 2024
Quantification of gDNA
Used Lane lab nanodrop and kit elution buffer as blank to quantify the first column of DNA on the plate.
| Sample ID | Well | Project | Collection Date | ng/µl | A260/280 | A260/230 |
|---|---|---|---|---|---|---|
| POC-466 | A1 | POC HISTOLOGY | Sept 2022 | 5.1 | 2.14 | 0.97 |
| POC-478 | B1 | POC HISTOLOGY | Sept 2022 | 7.6 | 2.16 | 2.04 |
| FLOW-005 | C1 | FLOW SKELETON | March 2024 | 15.9 | 2.08 | 2.14 |
| FLOW-020 | D1 | FLOW SKELETON | March 2024 | 10.6 | 1.96 | 1.77 |
| FLOW-033 | E1 | FLOW SKELETON | March 2024 | 6.8 | 2.40 | 0.85 |
| TPC REC-001 | F1 | TPC COTS RAPID | March 2024 | 12.3 | 2.00 | 1.89 |
| TPC REC-013 | G1 | TPC COTS RAPID | March 2024 | 10.3 | 2.04 | 1.19 |
| TPC REC-025 | H1 | TPC COTS RAPID | March 2024 | 22.8 | 1.98 | 1.58 |
| Elution Buffer | NA | NA | NA | 0.1 | -0.46 | 0.36 |
PCR
Prepare Reagents
Primers - Flot et al. 2008
- Forward primer FatP6.1 (Added 50µl of stock 2mg/ml plus 950µl of Nuclease Free water and made 3 aliquots of ~320µl each and stored in reagent box at -20)
- FatP6.1 (5′-TTTGGGSATTCGTTTAGCAG-3′)
- Reverse primer RORF (Added 50µl of stock 2mg/ml plus 950µl of Nuclease Free water and made 3 aliquots of ~320µl each and stored in reagent box at -20)
- RORF (5′-SCCAATATGTTAAACASCATGTCA-3′)
- Master Mix EmeraldAmp GT PCR Master Mix
EmeraldAmp GT PCR Master Mix is a loading-dye-added version of EmeraldAmp MAX PCR Master Mix that is optimized for great performance and convenience in both standard and high-throughput PCR applications.EmeraldAmp GT PCR Master Mix was recieved warm on arrival and stored at -20°C
Master Mix: For 82 rxns +3 = 85
| Reagent | 1Rxn µl | 85 Rxn µl |
|---|---|---|
| EmeraldAmp GT Mix (2x) | 12.5 | 1062.5 |
| F primer FatP6.1 (10µM) | 0.3 | 25.5 |
| R primer RORF (10µM) | 0.3 | 25.5 |
| DNA | 1 | NA |
| H2O | 10.9 | 926.5 |
| Total volume | 25 | —- |
24µl added to each of 82 wells. The total master mix was not enough and 2 more wells needed to be generated individually (H8 and H9). Next time add more for pipette loss.
Thermal Cycling Conditions
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 75 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
PCR Gel protocol
DNA was assessed with a 1.5% agarose gel in 1x TAE buffer (2.25g agarose in 150ml of 1X TAE, Tris base, acetic acid and EDTA) run at for 40 mins at 66 V and stained with Biotium GelGreen Nucleic Acid Gel Stain (10,000X in Water Fisher Cat NC9728313).
Gel Results

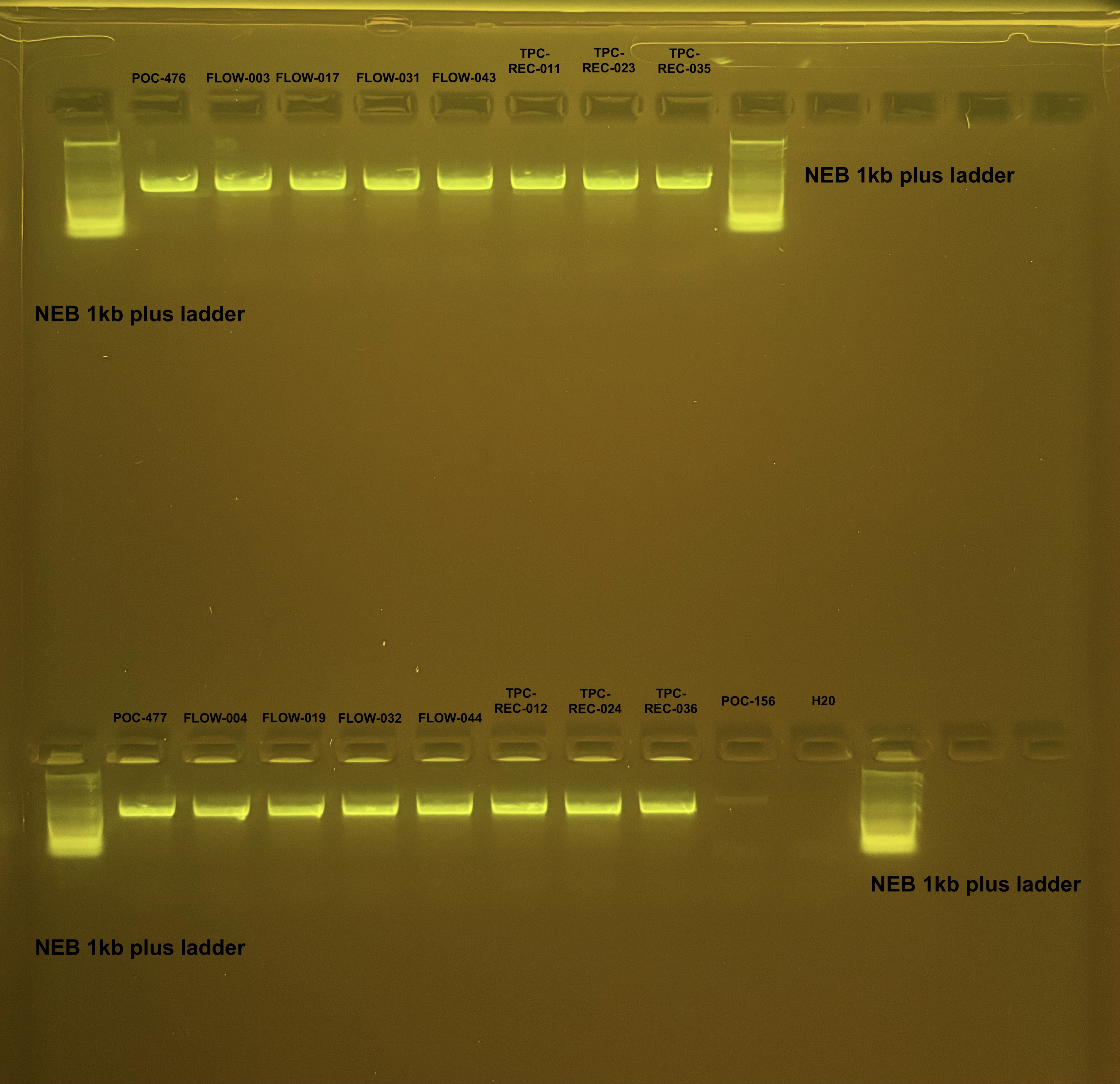
Pocillopora mtORF PCR and Pocillopora gDNA Gel results 4/12/24
Bands of ~1000 bp are present in all samples except the negative control for the mtORF amplicons.
Bands of HMW were present for all of the gDNA.
Amplified samples were saved in PCR plate for cleanup for mtORF For primer Sanger sequencing in 4°C fridge by PCR bench.
20240419
DNA extraction of remaining 16 samples from Plate001
DNA was extracted withZymo Research Quick-DNA™ Miniprep plus kit extraction Cat D4068
ProteinaseK was already added from steps above, so started at step 3 on page 7.
- Added 300µl of Genomic Binding Buffer to the sample and mixed with the pipette.
- Transferred the mixture to a Zymo-Spin™ IIC-XLR Column in a Collection Tube. Centrifuged at 12,000 x g for 1 minute. Discarded the flow through and collection tube.
- Added 400 µl DNA Pre-Wash Buffer to the spin column in a new Collection Tube. Centrifuged at 12,000 x g for 1 minute. Emptied the Collection Tube.
- Added 700 µl g-DNA Wash Buffer to the spin column. Centrifuged at 12,000 x g for 1 minute. Emptied the Collection Tube.
- Added 200 µl g-DNA Wash Buffer to the spin column. Centrifuged at 12,000 x g for 1 minute. Discarded the Collection Tube with the flow through.
- TransferRED the spin column to a clean microcentrifuge tube. Add 60 µl DNA Elution Buffer that was heated to 70°C and incubated for 5 minutes at room temperature, then centrifuged at 20,000 x g for 1 minute to elute the DNA.
- The eluted DNA was then quantified via nanodrop, used for PCR, and stored -20ºC in box 1 of a new vertical metal rack in chest freezer for future use.
- FLOW-003 lid broke in spin
- FLOW-043 lid cracked in spin
Quantification of gDNA
Used Lane lab nanodrop and kit elution buffer as blank to quantify the 16 samples.
| Sample ID | Well | Project | Collection Date | ng/µl | A260/280 | A260/230 |
|---|---|---|---|---|---|---|
| POC-476 | A11 | POC HISTOLOGY | Sept 2022 | 14.8 | 2.10 | 0.63 |
| FLOW-003 | C11 | FLOW SKELETON | March 2024 | 38.4 | 1.91 | 1.97 |
| FLOW-017 | C11 | FLOW SKELETON | March 2024 | 28.7 | 2.01 | 0.11 |
| FLOW-031 | D11 | FLOW SKELETON | March 2024 | 42.9 | 1.97 | 1.96 |
| FLOW-043 | E11 | FLOW SKELETON | March 2024 | 45.1 | 1.94 | 1.88 |
| TPC REC-011 | F11 | TPC COTS RAPID | March 2024 | 11.1 | 2.20 | 0.04 |
| TPC REC-023 | G11 | TPC COTS RAPID | March 2024 | 94.0 | 1.92 | 0.38 |
| TPC REC-035 | H11 | TPC COTS RAPID | March 2024 | 2.2 | 3.04 | 0.19 |
| POC-477 | A11 | POC HISTOLOGY | Sept 2022 | 15.7 | 2.00 | 1.44 |
| FLOW-004 | C11 | FLOW SKELETON | March 2024 | 55.3 | 1.93 | 1.93 |
| FLOW-019 | C11 | FLOW SKELETON | March 2024 | 38.4 | 1.97 | 2.11 |
| FLOW-032 | D11 | FLOW SKELETON | March 2024 | 46.3 | 1.86 | 1.54 |
| FLOW-044 | E11 | FLOW SKELETON | March 2024 | 20.5 | 1.99 | 1.00 |
| TPC REC-012 | F11 | TPC COTS RAPID | March 2024 | 15.0 | 2.10 | 1.58 |
| TPC REC-024 | G11 | TPC COTS RAPID | March 2024 | 58.2 | 1.93 | 2.39 |
| TPC REC-036 | H11 | TPC COTS RAPID | March 2024 | 6.8 | 2.66 | 0.84 |
| Elution Buffer | H11 | TPC COTS RAPID | March 2024 | 0.5 | -1.24 | 0.39 |
Master Mix: For 18 rxns +2 = 20
| Reagent | 1Rxn µl | 20 Rxn µl |
|---|---|---|
| EmeraldAmp GT Mix (2x) | 12.5 | 250 |
| F primer FatP6.1 (10µM) | 0.3 | 6 |
| R primer RORF (10µM) | 0.3 | 6 |
| DNA | 1 | NA |
| H2O | 10.9 | 218 |
| Total volume | 25 | —- |
24µl of master mix was added to each of 18 wells positive control = POC 156 Spawning negative control = master mix water
Thermal Cycling Conditions
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 75 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
PCR Gel protocol
DNA was assessed with a small 28 well 1.5% agarose gel in 1x TAE buffer (1.12g agarose in 75ml of 1X TAE, Tris base, acetic acid and EDTA) run at for 20 mins at 100 V and stained with 1.5 µl of Biotium GelGreen Nucleic Acid Gel Stain (10,000X in Water Fisher Cat NC9728313). 4 µl of sample or 4µl of NEB 1kb plus purple ladder were added to each well.
Gel Results
PCR bands of the expected size were seen in all lanes except the negative control. PCR product was stored in the 4°C fridge by the PCR bench with the rest of Plate 001.
20240421
PCR Cleanup
1X KapaPure Bead Cleanup (All work was conducted on post PCR bench)
KAPAPure beads are light sensitive
KapaPure bead 1x KapaPure bead Tech Sheet
Take KAPA pure beads out of 4 degree fridge ~30 minutes before use to get to room temperature, and swirl to even out the beads (which settle on the bottom), and keep them in a drawer while warming out of the light
-
Made fresh 80% ethanol by adding 32ml 100% ethanol and 8ml of nucelase free water for 40mL total
-
Took the mtORF PCR plate001 and 16 strip tubes out of the fridge and brought to room temperature
-
Added 2ml of KapaPure beads to a sterile trough and used the multichannel pipette to add 20ul of beads to each of 96 wells and pipette up and down to mix and bind DNA to the beads
-
Placed the sample plate on the rotating shaker to incubate at RT for 10 minutes at 100 speed to facilitate DNA binding.
-
After incubation, placed the plate on the magnet for 1min until the solution was clear and removed the supernatant from every well into the waste trough
-
Added 200ul of freshly made 80% EtOH to each well carefully without disturbing the beads.
-
Removed the clear supernatant from each well without disturbing the beads and discard into the waste trough.
-
Repeated the 80% ethanol washes and removals for a total of 2 times.
-
While the plate is on the magnet used the p20 multichannel pipette to remove any remaining ethanol at the bottom of the wells of the plate
-
Waited ~3-5 minutes to let any residual ethanol to dry and took the plate off the magnet into a regular rack.
Do not overdry the beads
-
Added 40ul of Zymo Elution Buffer to each well of the plate and mixed beads into resuspension by pipetting.
-
Removed 35ul of clear supernatant from each well and into a new 96-well plate
-
set up a Sanger sequencing run below and then sealed the plate with a labeled foil seal and placed it in the fridge.
Sanger sequencing setup
-
Diluted FatP6.1 Forward primer to 3.2 µM (100µL 10µM primer + 200µL of nucelase free water)
-
Added 8µl of nucease free water, 2ul from each sample, and 2µL of FatP6.1 Forward primer (at 3.2 µM) into a new plate and covered with a foil seal.
-
Labelled the seal with POC COTS RAPID Putnam 20240421 HP0001 - HP0096
-
Sanger sequenced received as folders HP_240423B and HP_240425 and stored in respective github repos.
