Poc ID via mtORF
Pocillopora species ID for POC ID relative abundance and Pocillopora spawning Moorea 2022
Equipment and Reagents
- Sample Preservation and Lysis Zymo Research DNA/RNA Shield R1100-250
- DNA Extraction Zymo Research Quick-DNA™ Miniprep Plus Kit D4069
- Foward primer FatP6.1 200µM Stock IDT)
- Reverse primer RORF 200µM Stock IDT
- Master Mix EmeraldAmp GT PCR Master Mix
- EmeraldAmp GT PCR Master Mix is a loading-dye-added version of EmeraldAmp MAX PCR Master Mix that is optimized for great performance and convenience in both standard and high-throughput PCR applications.
- Loading Dye NEB 6X Purple Loading Dye NEB Cat # B7024S
- Gel Stain Biotium GelGreen Nucleic Acid Gel Stain, 10,000X in Water Fisher Cat NC9728313
- DNA Ladder 1kb Gel ladder
- Gel ladder GeneRuler 100 bp DNA Ladder (Thermo FIsher Catalog SM0241)
- Restriction Enzyme XhoI R0146Sk
- Restriction Enzyme Buffer rCutSmart B6004S
23 July 2023
DNA Extraction
DNA Extraction Zymo Research Quick-DNA™ Miniprep Plus Kit D4069
DNA was extracted with Quick-DNA™ Miniprep Plus Kit (Zymo Research Cat # D4069) First 200µl of DNA/RNA Shield (Zymo Research Cat # R1100-250) containing the lysed sample was moved to a new microfuge tube and mixed with 200µl of new DNA/RNA Shield. The samples were extracted according to the manufacturer’s instructions for samples stored in DNA/RNA Shield including the addition of 20µl of Proteinase K (20mg ml-1). Samples were eluted in 50µl of kit Elution Buffer warmed to 65°C and DNA was assessed with a 1.5% agarose gel in 1x TAE buffer (Tris base, acetic acid and EDTA) run at for 45 mins at 100 V and stained with Biotium GelGreen Nucleic Acid Gel Stain (10,000X in Water Fisher Cat NC9728313).
DNA extractions were completed on 23 July 2023 and DNA stored at -20°C
Samples
| Sample ID | Project | Collection Date |
|---|---|---|
| NEB 1kb DNA Ladder | NEB N3232S | NA |
| POC-152 | Moorea TPC Relative Abundance | May 2022 |
| POC-155 | Moorea TPC Relative Abundance | May 2022 |
| POC-157 | Moorea TPC Relative Abundance | May 2022 |
| POC-160 | Moorea TPC Relative Abundance | May 2022 |
| POC-163 | Moorea TPC Relative Abundance | May 2022 |
| POC-164 | Moorea TPC Relative Abundance | May 2022 |
| POC-170 | Moorea TPC Relative Abundance | May 2022 |
| POC-171 | Moorea TPC Relative Abundance | May 2022 |
| POC-172 | Moorea TPC Relative Abundance | May 2022 |
| POC-173 | Moorea TPC Relative Abundance | May 2022 |
| POC-182 | Moorea TPC Relative Abundance | May 2022 |
| POC-131 | Moorea Poc Spawning | December 2022 |
| POC-132 | Moorea Poc Spawning | December 2022 |
| POC-133 | Moorea Poc Spawning | December 2022 |
| POC-134 | Moorea Poc Spawning | December 2022 |
| POC-135 | Moorea Poc Spawning | December 2022 |
| POC-176 | Moorea Poc Spawning | December 2022 |
| POC-185 | Moorea Poc Spawning | December 2022 |
| POC-186 | Moorea Poc Spawning | December 2022 |
| POC-189 | Moorea Poc Spawning | December 2022 |
| POC-191 | Moorea Poc Spawning | December 2022 |
| POC-194 | Moorea Poc Spawning | December 2022 |
| POC-195 | Moorea Poc Spawning | December 2022 |
| POC-196 | Moorea Poc Spawning | December 2022 |
| POC-198 | Moorea Poc Spawning | December 2022 |
| POC-199 | Moorea Poc Spawning | December 2022 |
| POC-200 | Moorea Poc Spawning | December 2022 |
| POC-578 | Moorea Poc Spawning | November 2022 |
| NEB 1kb DNA Ladder | NEB N3232S | NA |
24 July 2023
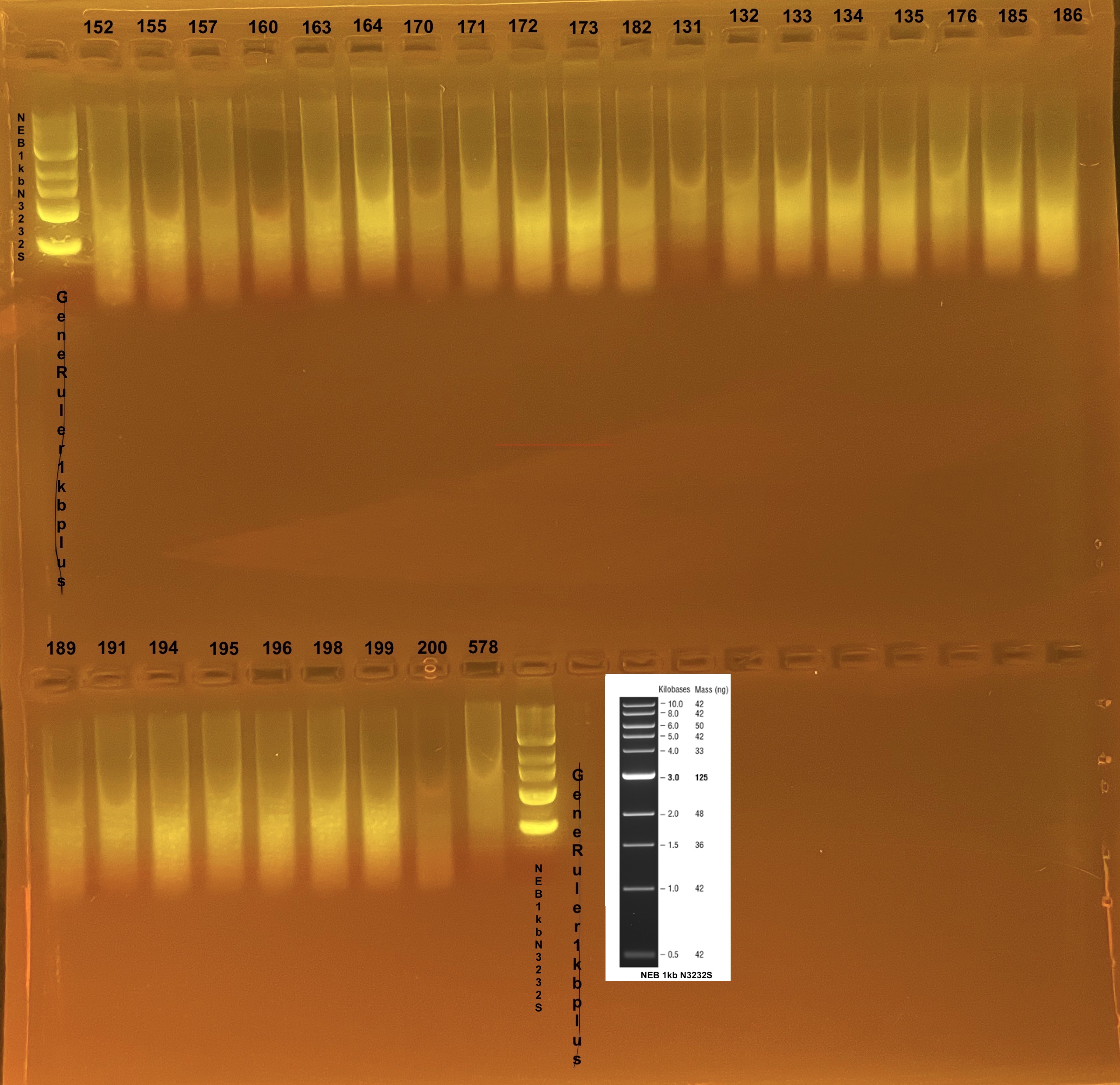
Gel QC of gDNA
Gel protocol
prepared a 100ml gel using 50ml of 1xTAE + 1.5g Agarose, melted gel for ~120 sec in the microwave and let cool and added 1µl Gelgreen. Ran gel at 100V for 45 minutes.
gDNA results
PCR
Prepare Reagents
Primers - Flot et al. 2008
- Forward primer FatP6.1 (Added 50µl of stock 2mg/ml plus 950µl of Nuclease Free water and made 3 aliquots of ~320µl each and stored in reagent box at -20)
- FatP6.1 (5′-TTTGGGSATTCGTTTAGCAG-3′)
- Reverse primer RORF (Added 50µl of stock 2mg/ml plus 950µl of Nuclease Free water and made 3 aliquots of ~320µl each and stored in reagent box at -20)
- RORF (5′-SCCAATATGTTAAACASCATGTCA-3′)
- Master Mix EmeraldAmp GT PCR Master Mix
EmeraldAmp GT PCR Master Mix is a loading-dye-added version of EmeraldAmp MAX PCR Master Mix that is optimized for great performance and convenience in both standard and high-throughput PCR applications.EmeraldAmp GT PCR Master Mix was recieved warm on arrival and stored at -20°C
EmeraldAmp GT PCR Master Mix (2x)
Master Mix: For 30 rxns +2 = 32
| Reagent | 1Rxn µl | 32 Rxn µl |
|---|---|---|
| EmeraldAmp GT Mix (2x) | 12.5 | 400 |
| F primer FatP6.1 (10µM) | 0.3 | 10.4 |
| R primer RORF (10µM) | 0.3 | 10.4 |
| DNA | 1 | NA |
| H2O | 10.9 | 347.2 |
| Total volume | 25 | 800 |
Thermal Cycling Conditions
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 75 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
PCR Gel protocol
prepared a 100ml gel using 50ml of 1xTAE + 1.5g Agarose, melted gel for ~120 sec in the microwave and let cool and added 1µl Gelgreen and let harden. Ran gel at 100V for 45 minutes in 1x TAE.
Gel Samples
Top Row
| Sample ID | Project | Collection Date |
|---|---|---|
| NEB 1kb DNA Ladder | NEB N3232S | NA |
| POC-152 | Moorea TPC Relative Abundance | May 2022 |
| POC-155 | Moorea TPC Relative Abundance | May 2022 |
| POC-157 | Moorea TPC Relative Abundance | May 2022 |
| POC-160 | Moorea TPC Relative Abundance | May 2022 |
| POC-163 | Moorea TPC Relative Abundance | May 2022 |
| POC-164 | Moorea TPC Relative Abundance | May 2022 |
| POC-170 | Moorea TPC Relative Abundance | May 2022 |
| POC-171 | Moorea TPC Relative Abundance | May 2022 |
| POC-172 | Moorea TPC Relative Abundance | May 2022 |
| POC-173 | Moorea TPC Relative Abundance | May 2022 |
| POC-182 | Moorea TPC Relative Abundance | May 2022 |
| POC-131 | Moorea Poc Spawning | December 2022 |
| POC-132 | Moorea Poc Spawning | December 2022 |
| POC-133 | Moorea Poc Spawning | December 2022 |
| POC-134 | Moorea Poc Spawning | December 2022 |
| POC-135 | Moorea Poc Spawning | December 2022 |
| POC-176 | Moorea Poc Spawning | December 2022 |
| POC-185 | Moorea Poc Spawning | December 2022 |
| NEB 1kb DNA Ladder | NEB N3232S | NA |
Bottom Row
| Sample ID | Project | Collection Date |
|---|---|---|
| NEB 1kb DNA Ladder | NEB N3232S | NA |
| POC-186 | Moorea Poc Spawning | December 2022 |
| POC-189 | Moorea Poc Spawning | December 2022 |
| POC-191 | Moorea Poc Spawning | December 2022 |
| POC-194 | Moorea Poc Spawning | December 2022 |
| POC-195 | Moorea Poc Spawning | December 2022 |
| POC-196 | Moorea Poc Spawning | December 2022 |
| POC-198 | Moorea Poc Spawning | December 2022 |
| POC-199 | Moorea Poc Spawning | December 2022 |
| POC-200 | Moorea Poc Spawning | December 2022 |
| POC-578 | Moorea Poc Spawning | November 2022 |
| Positive Control | H2 Hollie 2 chelex extraction of P. acuta 4/9/22 | 4/9/22 |
| Negative Control | Master Mix H20 | NA |
| NEB 1kb DNA Ladder | NEB N3232S | NA |
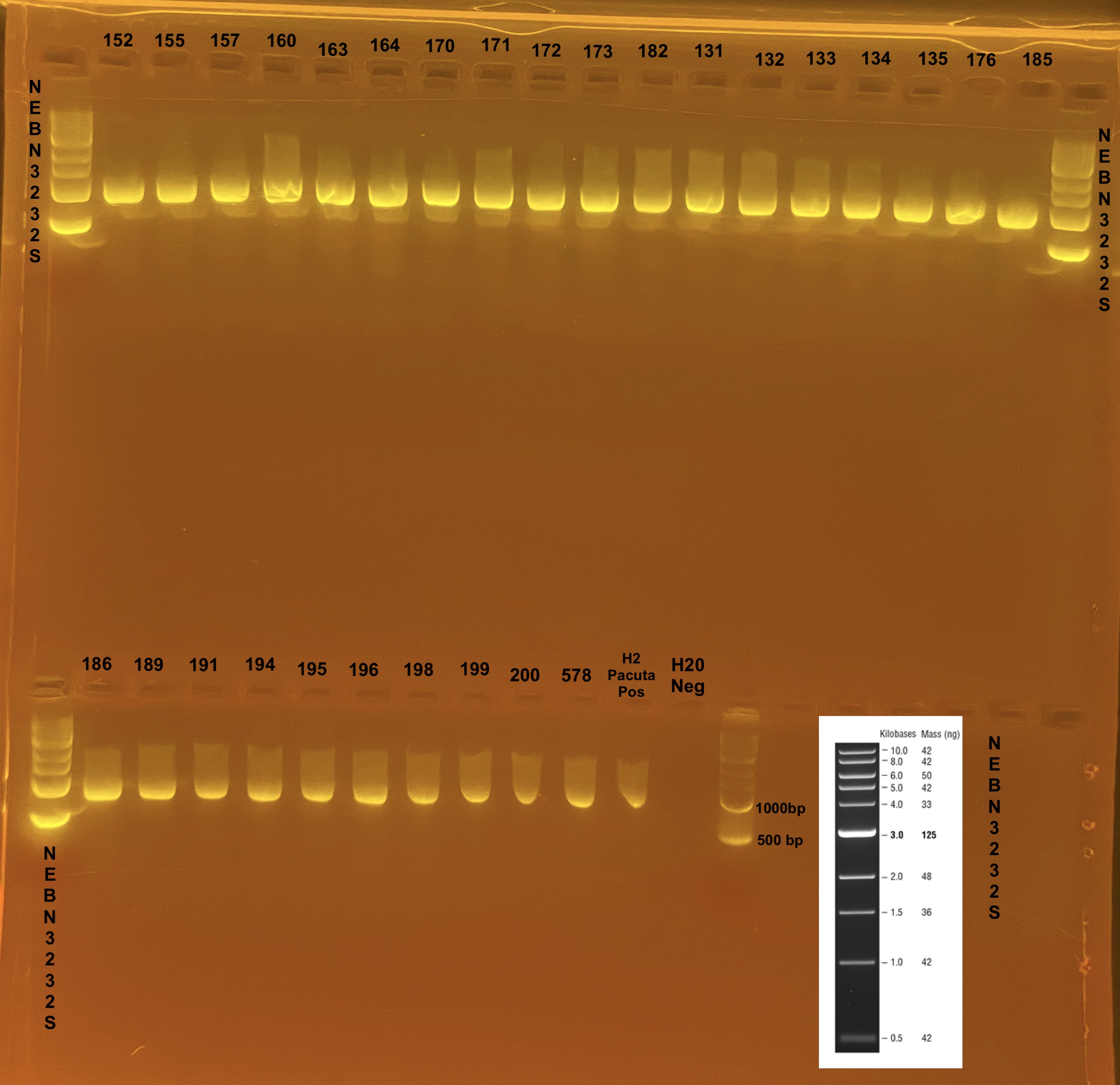
Gel Results
Bands of ~1000 bp are present in all samples except the negative control
PCR Cleanup
PCR Purification Qiagen MinElute
Eluted in 10 µl of EB. Elution repeated in another 10µl of EB, for a total of 20µl of purified PCR product.
#Sanger Sequencing Prep URI MIC Maggie Schedl Notebook
Qubit DNA quantification
Qubit™ dsDNA BR Assay Kit Catalog Numbers Q32850, Q32853
| Sample ID | Reading1 ng/µl | Reading1 ng/µl | Avg ng/µl |
|---|---|---|---|
| POC-152 | 31.2 | 31.0 | 31.1 |
| POC-155 | 33.8 | 33.8 | 33.8 |
| POC-157 | 31.0 | 30.6 | 30.8 |
| POC-160 | 30.0 | 29.8 | 29.9 |
| POC-163 | 38.2 | 38.0 | 38.1 |
| POC-164 | 30.6 | 31.2 | 30.9 |
| POC-170 | 30.8 | 30.6 | 30.7 |
| POC-171 | 27.4 | 27.4 | 27.4 |
| POC-172 | 30.8 | 30.8 | 30.8 |
| POC-173 | 35.8 | 35.8 | 35.8 |
| POC-182 | 31.2 | 31.0 | 31.1 |
| POC-131 | 31.2 | 31.2 | 31.2 |
| POC-132 | 35.6 | 35.4 | 35.5 |
| POC-133 | 30.2 | 30.2 | 30.2 |
| POC-134 | 32.8 | 33.6 | 33.2 |
| POC-135 | 34.2 | 34.2 | 34.2 |
| POC-176 | 32.0 | 32.0 | 32.0 |
| POC-185 | 31.8 | 32.0 | 31.9 |
| POC-186 | 33.8 | 33.6 | 33.7 |
| POC-189 | 29.2 | 29.2 | 29.2 |
| POC-191 | 28.6 | 28.4 | 28.5 |
| POC-194 | 38.4 | 38.2 | 38.3 |
| POC-195 | 38.4 | 38.2 | 38.3 |
| POC-196 | 39.6 | 39.4 | 39.5 |
| POC-198 | 32.2 | 32.2 | 32.2 |
| POC-199 | 22.4 | 22.4 | 22.4 |
| POC-200 | 28.8 | 28.8 | 28.8 |
| POC-578 | 33.0 | 33.0 | 33.0 |
Sample Dilution for For and Rev plates
Dilution of Template
2µl of PCR product + 8µl of H2O
Dilution of Primer
32µl Primer + 68µl H20 = 100µl of 3.2µM primer
Sample Submission Prep
10µl of Diluted template + 2µl of 3.2µM primer = 12µl submission
Samples were prepared in 8 tube strips with HP1 - HP56 labels
| Sequencing ID | Sample ID | Primer |
|---|---|---|
| HP1 | POC-152 | Forward Primer FatP6.1 |
| HP2 | POC-155 | Forward Primer FatP6.1 |
| HP3 | POC-157 | Forward Primer FatP6.1 |
| HP4 | POC-160 | Forward Primer FatP6.1 |
| HP5 | POC-163 | Forward Primer FatP6.1 |
| HP6 | POC-164 | Forward Primer FatP6.1 |
| HP7 | POC-170 | Forward Primer FatP6.1 |
| HP8 | POC-171 | Forward Primer FatP6.1 |
| HP9 | POC-172 | Forward Primer FatP6.1 |
| HP10 | POC-173 | Forward Primer FatP6.1 |
| HP11 | POC-182 | Forward Primer FatP6.1 |
| HP12 | POC-131 | Forward Primer FatP6.1 |
| HP13 | POC-132 | Forward Primer FatP6.1 |
| HP14 | POC-133 | Forward Primer FatP6.1 |
| HP15 | POC-134 | Forward Primer FatP6.1 |
| HP16 | POC-135 | Forward Primer FatP6.1 |
| HP17 | POC-176 | Forward Primer FatP6.1 |
| HP18 | POC-185 | Forward Primer FatP6.1 |
| HP19 | POC-186 | Forward Primer FatP6.1 |
| HP20 | POC-189 | Forward Primer FatP6.1 |
| HP21 | POC-191 | Forward Primer FatP6.1 |
| HP22 | POC-194 | Forward Primer FatP6.1 |
| HP23 | POC-195 | Forward Primer FatP6.1 |
| HP24 | POC-196 | Forward Primer FatP6.1 |
| HP25 | POC-198 | Forward Primer FatP6.1 |
| HP26 | POC-199 | Forward Primer FatP6.1 |
| HP27 | POC-200 | Forward Primer FatP6.1 |
| HP28 | POC-578 | Forward Primer FatP6.1 |
| HP29 | POC-152 | Reverse Primer RORF |
| HP30 | POC-155 | Reverse Primer RORF |
| HP31 | POC-157 | Reverse Primer RORF |
| HP32 | POC-160 | Reverse Primer RORF |
| HP33 | POC-163 | Reverse Primer RORF |
| HP34 | POC-164 | Reverse Primer RORF |
| HP35 | POC-170 | Reverse Primer RORF |
| HP36 | POC-171 | Reverse Primer RORF |
| HP37 | POC-172 | Reverse Primer RORF |
| HP38 | POC-173 | Reverse Primer RORF |
| HP39 | POC-182 | Reverse Primer RORF |
| HP40 | POC-131 | Reverse Primer RORF |
| HP41 | POC-132 | Reverse Primer RORF |
| HP42 | POC-133 | Reverse Primer RORF |
| HP43 | POC-134 | Reverse Primer RORF |
| HP44 | POC-135 | Reverse Primer RORF |
| HP45 | POC-176 | Reverse Primer RORF |
| HP46 | POC-185 | Reverse Primer RORF |
| HP47 | POC-186 | Reverse Primer RORF |
| HP48 | POC-189 | Reverse Primer RORF |
| HP49 | POC-191 | Reverse Primer RORF |
| HP50 | POC-194 | Reverse Primer RORF |
| HP51 | POC-195 | Reverse Primer RORF |
| HP52 | POC-196 | Reverse Primer RORF |
| HP53 | POC-198 | Reverse Primer RORF |
| HP54 | POC-199 | Reverse Primer RORF |
| HP55 | POC-200 | Reverse Primer RORF |
| HP56 | POC-578 | Reverse Primer RORF |
Submission
Submitted samples to URI MIC for Sanger Seq on 7/26/23 MIC will sequence the first 4 Forward and the first 4 reverse to check sequencing quality before full run.
All sequences were completed in Forward and Reverse
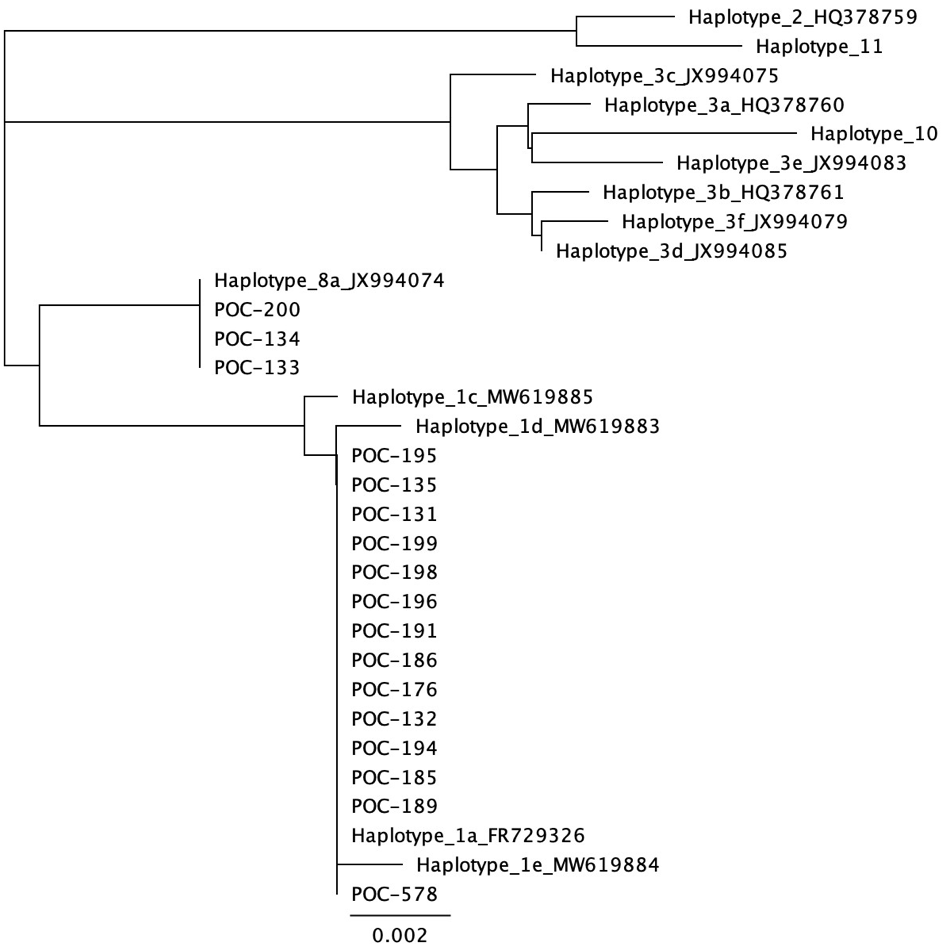
Geneious Prime analysis of mtORF
Analysis of data indicated 3 samples were Haplotype 8a and 14 samples were Haplotype 1a, which could be Pocillopora meandrina or Pocillopora grandis. PocHistone PCR and RFLP was used to identify the species for Haplotype 1a according to Johnston et al. 2018
Raw seqeunce data, reference sequences, and alignments
PocHistone PCR
15 August 2023
Prepare Reagents
Primers - Johnston et al. 2018
- Forward primer (PocHistoneF: 5′-ATTCAGTCTCACTCACTCACTCAC- 3′)
- Reverse primer (PocHistoneR: 5′-TATCTTCGAACAGACCCACCAAAT-3′)
- Master Mix EmeraldAmp GT PCR Master Mix
Primers were resuspended to 100µM and diluted to 10µM for use in PCR.
EmeraldAmp GT PCR Master Mix (2x)
Master Mix: For 16 rxns +2 = 18
| Reagent | 1Rxn µl | 18 Rxn µl |
|---|---|---|
| EmeraldAmp GT Mix (2x) | 12.5 | 225 |
| F primer PocHistoneF (10µM) (10µM) | 0.3 | 5.4 |
| R primer PocHistoneR (10µM) (10µM) | 0.3 | 5.4 |
| DNA | 1 | NA |
| H2O | 10.9 | 196.2 |
| Total volume | 25 | 450 |
Thermal Cycling Conditions
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 60 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
- stored at 4°C overnight at 19:54
PocHistone PCR Gel and RFLP
15 August 2023 Restriction Digest
Ran PocHIstone RFLP with NEB Restriction Enzyme XhoI, NEB Cat# R0146S
- Restriction Enzyme XhoI R0146Sk
- Restriction Enzyme Buffer rCutSmart B6004S
Restriction Digest Master Mix
Reagent |1Rxn µl |20 Rxn µl|
—|—|—|
XhoI restriction enzyme | 0.5 |10 |
1X CutSmartR buffer |0.5| 10 |
For each sample added 1µl of Restriction Digest Master Mix to 15µl of PCR product. Incubated samples for 1 hr at 37°C followed by 20 min at 65°C to inactivate the enzymes.
PCR Gel protocol
prepared a 100ml gel using 100ml of 1xTAE + 2g Agarose, melted gel for ~120 sec in the microwave and let cool and added 1µl Gelgreen and let harden. Ran gel at 70V for 110 minutes in 1x TAE.
Gel Samples
Top Row and Bottom Row Sample order Top Row = PCR product Bottom Row = RFLP Product
| Sample ID | Project | Collection Date |
|---|---|---|
| GeneRuler 100 bp DNA Ladder | Thermo FIsher Cat SM0241 | NA |
| POC-131 | Moorea Poc Spawning | December 2022 |
| POC-132 | Moorea Poc Spawning | December 2022 |
| POC-135 | Moorea Poc Spawning | December 2022 |
| POC-176 | Moorea Poc Spawning | December 2022 |
| POC-185 | Moorea Poc Spawning | December 2022 |
| POC-186 | Moorea Poc Spawning | December 2022 |
| POC-189 | Moorea Poc Spawning | December 2022 |
| POC-191 | Moorea Poc Spawning | December 2022 |
| POC-194 | Moorea Poc Spawning | December 2022 |
| POC-195 | Moorea Poc Spawning | December 2022 |
| POC-196 | Moorea Poc Spawning | December 2022 |
| POC-198 | Moorea Poc Spawning | December 2022 |
| POC-199 | Moorea Poc Spawning | December 2022 |
| POC-578 | Moorea Poc Spawning | December 2022 |
| H2 | P acuta lab sample | 4/9/22 |
| Neg Control H20 | NA | NA |
| GeneRuler 100 bp DNA Ladder | Thermo FIsher Cat SM0241 | NA |

Gel Results
Top Row
For the PocHistone PCR, Bands of 669bp are present as expected with these primers in all samples except the negative control and the H2 P. acuta sample
Bottom Row
For the PocHistone PCR, Bands of 669bp are present, as expected with these primers if the samples are P. meandrina. Two bands at 287 and 382 are found in one sample POC-198, which indicates it is P. grandis (P. eydouxi). No bands were present in the negative control and the H2 P. acuta sample had the same two larger bands as in the PCR gel top row, as it is not cut by XhoI.
13 samples are P. meandrina and 1 sample is P. grandis.
P. meandrina
POC-131
POC-132
POC-135
POC-176
POC-185
POC-186
POC-189
POC-191
POC-194
POC-195
POC-196
POC-199
POC-578
P. grandis
POC-198
