Geoduck RRBS Library Prep - part 3
Prepping 40 RRBS libraries for geoduck juvenile OA acclimatization study.
20161214 100ng was used as the starting amount of DNA for each sample and the volume of water in each reaction was adjusted as necessary. 0.5% unmethylated lambda DNA was spiked in (w/w DNA) to determine conversion efficiency by estimating the error rate at which a C count occurs at an unmethylated C position.
Step 1 MSPI DNA Cutting
MSPI restriction endonuclease in NEBuffer2
- MspI - 25,000U (NEB cat: R0106L)
- NEBuffer2 10x (NEB cat: B7002S)
Reaction Mix
- Samples were mixed as follows to a total reaction volume of 30µl. Samples were incubated at 37°C starting at 18:00 on 20161214
- Lambda DNA (581ng/µl) was diluted 1:1160 for a concentration of 0.5ng/µl and 1µl added to each sample
-
Unmethylated lamda phage DNA (Promega cat: D1501)
- 3µl NEBuffer2
- 1µl MSPI 20U/µl
- 16µl DNA (100ng)
- 10µl of H20
- 30µl total
| Date | EPI.Tube.Num | Tank | Treatment | TimePoint | Homogenization | DNA.Extraction | DNA.QC | DNA.Conc.ng.µl | DNA.vol.µl | DNA.amount.µg | vol for 100ng | lambda.0.5% | Water.µl | NSBuffer2.µl | MSPI.µl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16-Mar-16 | EPI_41 | Time0 | Field | Day0 | 20160912 | 20161025 | 20161026 | 13.6 | 140 | 1.904 | 7.4 | 1 | 17.6 | 3 | 1 |
| 16-Mar-16 | EPI_42 | Time0 | Field | Day0 | 20160912 | 20161028 | 20161028 | 79 | 140 | 11.06 | 1.3 | 1 | 23.7 | 3 | 1 |
| 16-Mar-16 | EPI_43 | Time0 | Field | Day0 | 20160912 | 20161025 | 20161026 | 17.2 | 140 | 2.408 | 5.8 | 1 | 19.2 | 3 | 1 |
| 16-Mar-16 | EPI_44 | Time0 | Field | Day0 | 20160912 | 20161025 | 20161026 | 14.8 | 140 | 2.072 | 6.8 | 1 | 18.2 | 3 | 1 |
| 29-Jul-16 | EPI_151 | Ambient | Ambient | Day135 | 20160909 | 20161004 | 20161006 | 11.8 | 140 | 1.652 | 8.5 | 1 | 16.5 | 3 | 1 |
| 29-Jul-16 | EPI_152 | Ambient | Ambient | Day135 | 20160909 | 20161004 | 20161006 | 16.8 | 140 | 2.352 | 6 | 1 | 19 | 3 | 1 |
| 29-Jul-16 | EPI_153 | Ambient | Ambient | Day135 | 20160909 | 20161025 | 20161026 | 22.4 | 140 | 3.136 | 4.5 | 1 | 20.5 | 3 | 1 |
| 29-Jul-16 | EPI_154 | Ambient | Ambient | Day135 | 20160909 | 20161025 | 20161026 | 19.5 | 140 | 2.73 | 5.1 | 1 | 19.9 | 3 | 1 |
| 29-Jul-16 | EPI_159 | Low | Low | Day135 | 20160909 | 20161004 | 20161006 | 21.4 | 140 | 2.996 | 4.7 | 1 | 20.3 | 3 | 1 |
| 29-Jul-16 | EPI_160 | Low | Low | Day135 | 20160909 | 20161004 | 20161006 | 10.8 | 140 | 1.512 | 9.3 | 1 | 15.7 | 3 | 1 |
| 29-Jul-16 | EPI_161 | Low | Low | Day135 | 20160909 | 20161028 | 20161028 | 24.9 | 140 | 3.486 | 4 | 1 | 21 | 3 | 1 |
| 29-Jul-16 | EPI_162 | Low | Low | Day135 | 20160909 | 20161007 | 20161010 | 12.5 | 140 | 1.75 | 8 | 1 | 17 | 3 | 1 |
| 29-Jul-16 | EPI_167 | Super.Low | Super.Low | Day135 | 20160908 | 20160930 | Pass | 13.2 | 190 | 2.508 | 7.6 | 1 | 17.4 | 3 | 1 |
| 29-Jul-16 | EPI_168 | Super.Low | Super.Low | Day135 | 20160908 | 20160930 | Pass | 6.18 | 190 | 1.1742 | 16.2 | 1 | 8.8 | 3 | 1 |
| 29-Jul-16 | EPI_169 | Super.Low | Super.Low | Day135 | 20160908 | 20161007 | 20161010 | 14.8 | 140 | 2.072 | 6.8 | 1 | 18.2 | 3 | 1 |
| 29-Jul-16 | EPI_170 | Super.Low | Super.Low | Day135 | 20160908 | 20161007 | 20161010 | 11.2 | 140 | 1.568 | 8.9 | 1 | 16.1 | 3 | 1 |
| 8-Aug-16 | EPI_175 | Bin1 | Low-Ambient | Day145 | 20160908 | 20160930 | Pass | 19.6 | 190 | 3.724 | 5.1 | 1 | 19.9 | 3 | 1 |
| 8-Aug-16 | EPI_176 | Bin1 | Low-Ambient | Day145 | 20160908 | 20160930 | Pass | 7.84 | 190 | 1.4896 | 12.8 | 1 | 12.2 | 3 | 1 |
| 8-Aug-16 | EPI_181 | Bin2 | Ambient-Ambient | Day145 | 20160908 | 20161004 | 20161006 | 40.8 | 140 | 5.712 | 2.5 | 1 | 22.5 | 3 | 1 |
| 8-Aug-16 | EPI_182 | Bin2 | Ambient-Ambient | Day145 | 20160908 | 20161004 | 20161006 | 20.8 | 140 | 2.912 | 4.8 | 1 | 20.2 | 3 | 1 |
| 8-Aug-16 | EPI_184 | Bin3 | Ambient-Ambient | Day145 | 20160908 | 20161007 | 20161010 | 7.46 | 140 | 1.0444 | 13.4 | 1 | 11.6 | 3 | 1 |
| 8-Aug-16 | EPI_185 | Bin3 | Ambient-Ambient | Day145 | 20160907 | 20161007 | 20161010 | 18.6 | 140 | 2.604 | 5.4 | 1 | 19.6 | 3 | 1 |
| 8-Aug-16 | EPI_187 | Bin4 | Super.Low-Ambient | Day145 | 20160907 | 20160930 | Pass | 17.1 | 190 | 3.249 | 5.8 | 1 | 19.2 | 3 | 1 |
| 8-Aug-16 | EPI_188 | Bin4 | Super.Low-Ambient | Day145 | 20160907 | 20160930 | Pass | 14.5 | 190 | 2.755 | 6.9 | 1 | 18.1 | 3 | 1 |
| 8-Aug-16 | EPI_193 | Bin5 | Low-Ambient | Day145 | 20160907 | 20161004 | 20161006 | 9.26 | 140 | 1.2964 | 10.8 | 1 | 14.2 | 3 | 1 |
| 8-Aug-16 | EPI_194 | Bin5 | Low-Ambient | Day145 | 20160907 | 20161004 | 20161006 | 11.7 | 140 | 1.638 | 8.5 | 1 | 16.5 | 3 | 1 |
| 8-Aug-16 | EPI_199 | Bin6 | Super.Low-Ambient | Day145 | 20160907 | 20161004 | 20161006 | 21.8 | 140 | 3.052 | 4.6 | 1 | 20.4 | 3 | 1 |
| 8-Aug-16 | EPI_200 | Bin6 | Super.Low-Ambient | Day145 | 20160907 | 20161004 | 20161006 | 21.2 | 140 | 2.968 | 4.7 | 1 | 20.3 | 3 | 1 |
| 8-Aug-16 | EPI_205 | Bin7 | Ambient-Low | Day145 | 20160907 | 20160930 | Pass | 11.9 | 190 | 2.261 | 8.4 | 1 | 16.6 | 3 | 1 |
| 8-Aug-16 | EPI_206 | Bin7 | Ambient-Low | Day145 | 20160906 | 20160930 | Pass | 18.1 | 190 | 3.439 | 5.5 | 1 | 19.5 | 3 | 1 |
| 8-Aug-16 | EPI_208 | Bin8 | Low-Low | Day145 | 20160906 | 20161007 | 20161010 | 31 | 140 | 4.34 | 3.2 | 1 | 21.8 | 3 | 1 |
| 8-Aug-16 | EPI_209 | Bin8 | Low-Low | Day145 | 20160906 | 20161007 | 20161010 | 22 | 140 | 3.08 | 4.5 | 1 | 20.5 | 3 | 1 |
| 8-Aug-16 | EPI_214 | Bin9 | Super.Low-Low | Day145 | 20160906 | 20161004 | 20161006 | 13.3 | 140 | 1.862 | 7.5 | 1 | 17.5 | 3 | 1 |
| 8-Aug-16 | EPI_215 | Bin9 | Super.Low-Low | Day145 | 20160906 | 20161004 | 20161006 | 22.6 | 140 | 3.164 | 4.4 | 1 | 20.6 | 3 | 1 |
| 8-Aug-16 | EPI_220 | Bin10 | Super.Low-Low | Day145 | 20160906 | 20160930 | Pass | 48.8 | 190 | 9.272 | 2 | 1 | 23 | 3 | 1 |
| 8-Aug-16 | EPI_221 | Bin10 | Super.Low-Low | Day145 | 20160906 | 20160930 | Pass | 30 | 190 | 5.7 | 3.3 | 1 | 21.7 | 3 | 1 |
| 8-Aug-16 | EPI_226 | Bin11 | Ambient-Low | Day145 | 20160902 | 20161007 | 20161010 | 13.1 | 140 | 1.834 | 7.6 | 1 | 17.4 | 3 | 1 |
| 8-Aug-16 | EPI_227 | Bin11 | Ambient-Low | Day145 | 20160902 | 20161007 | 20161010 | 31.6 | 140 | 4.424 | 3.2 | 1 | 21.8 | 3 | 1 |
| 8-Aug-16 | EPI_229 | Bin12 | Low-Low | Day145 | 20160902 | 20160930 | Pass | 17.4 | 190 | 3.306 | 5.7 | 1 | 19.3 | 3 | 1 |
| 8-Aug-16 | EPI_230 | Bin12 | Low-Low | Day145 | 20160902 | 20160930 | Pass | 10.1 | 190 | 1.919 | 9.9 | 1 | 15.1 | 3 | 1 |
Step 1 conclusions
- no errors were seen on the PCR machine. Samples assumed to have been cut with MSPI
Step 2 Bisulfite conversion
- Removed from incubator at 09:00 on 20161215
- Proceeded with EZ DNA Methylation-Gold Kit (Catalog Nos. D5005 & D5006) EZ DNA Methylation-Gold Manual
Bisulfite conversion kit prep
-
Prepared the CT Conversion Reagent for larger DNA samples (30µl) by adding 800µl of water, 300µl of M-Dilution and 50µl of M-Dissolving
- Added 30µl of sample described in table to 120 µl of CT Conversion Reagent, mixed by flicking, and spun down
- Placed tubes in PCR machine at 10:45 and set as follows:
- 98°C for 10 min
- 64°C for 2.5 hours
- 4°C forever
-
followed protocol steps EZ DNA Methylation-Gold Manual
- Eluted in 12µl of elution buffer
Expected DNA size for Gold kit (A)
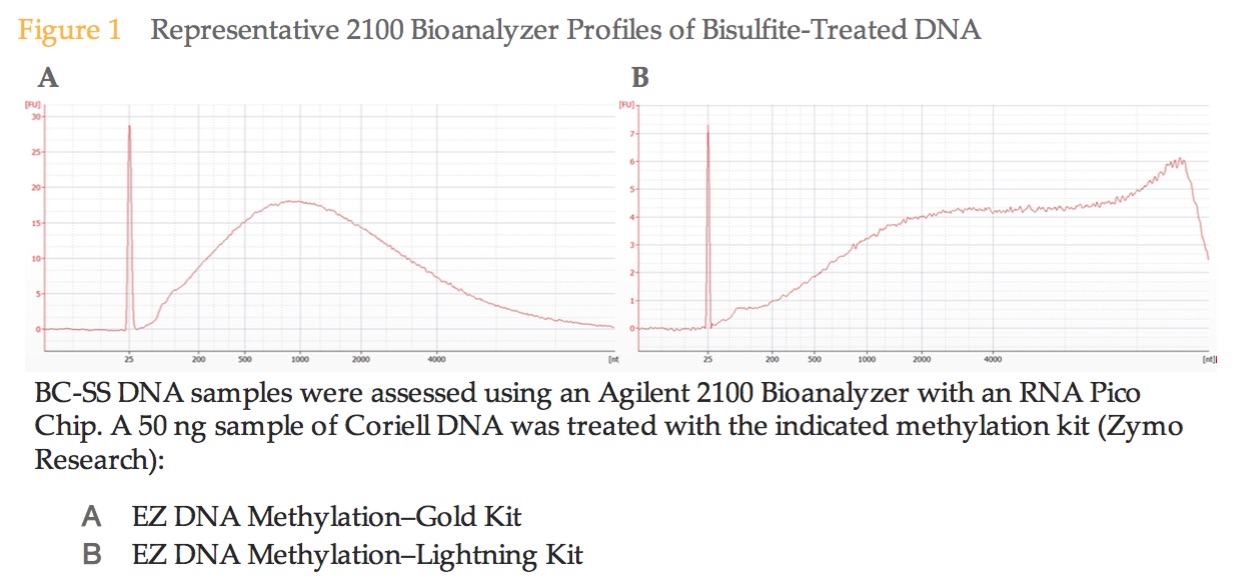
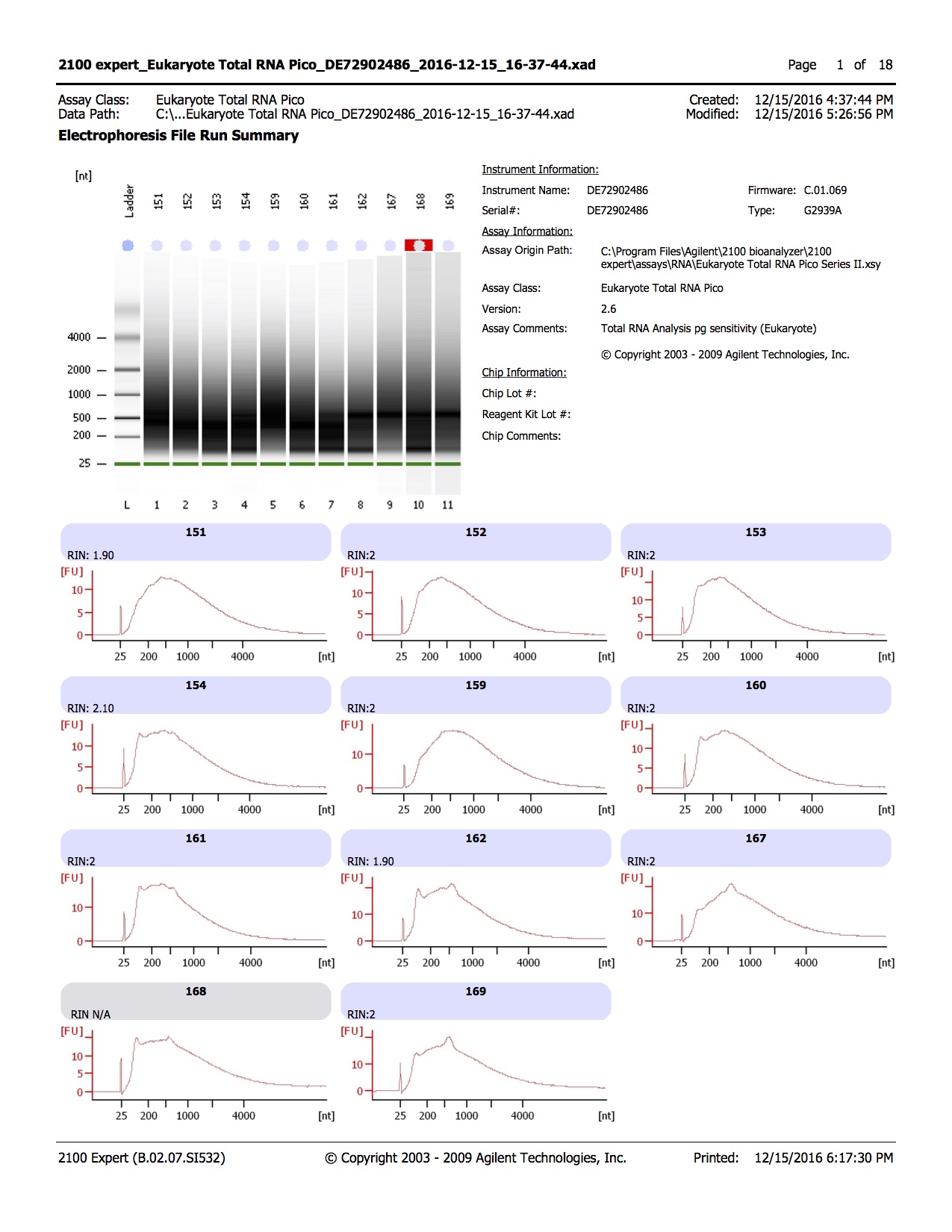
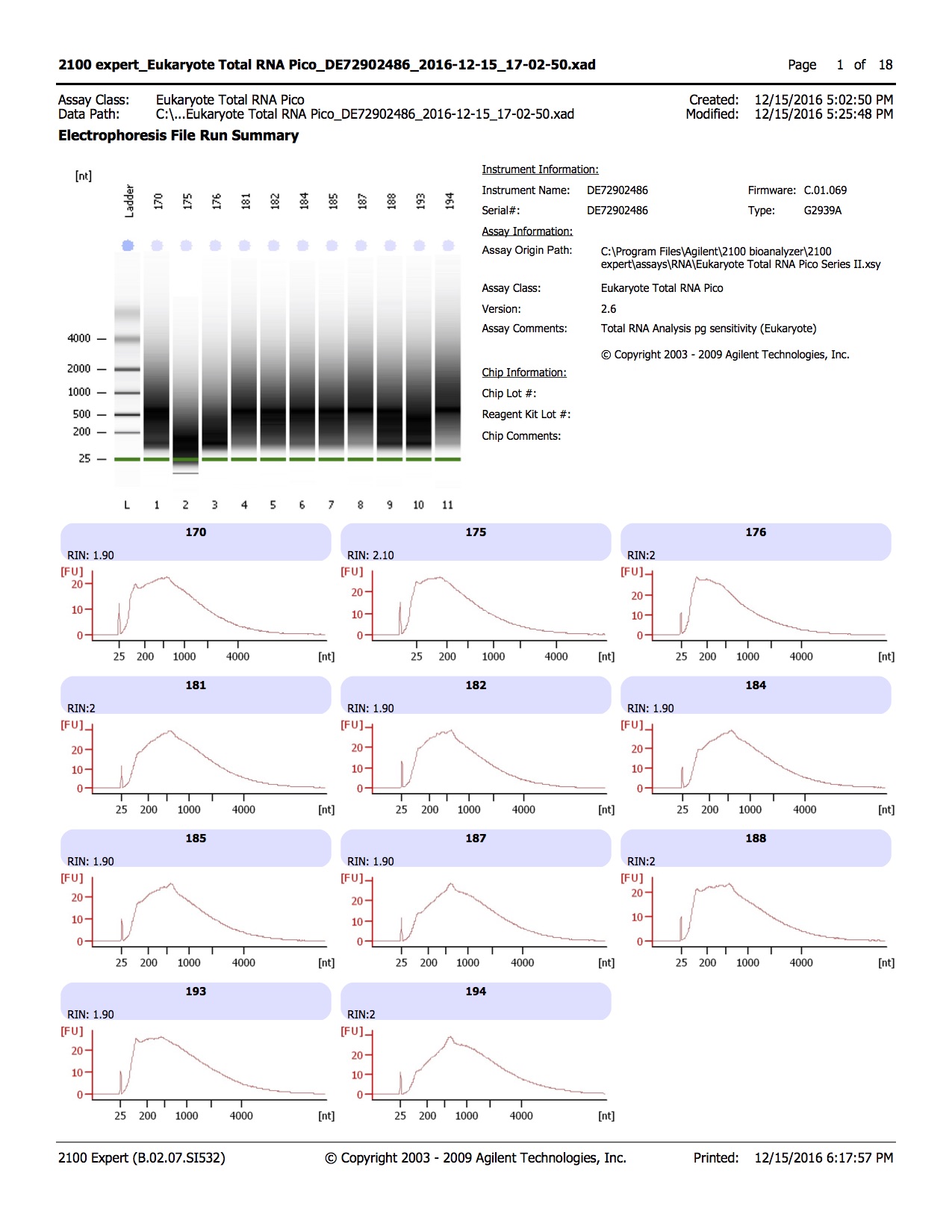
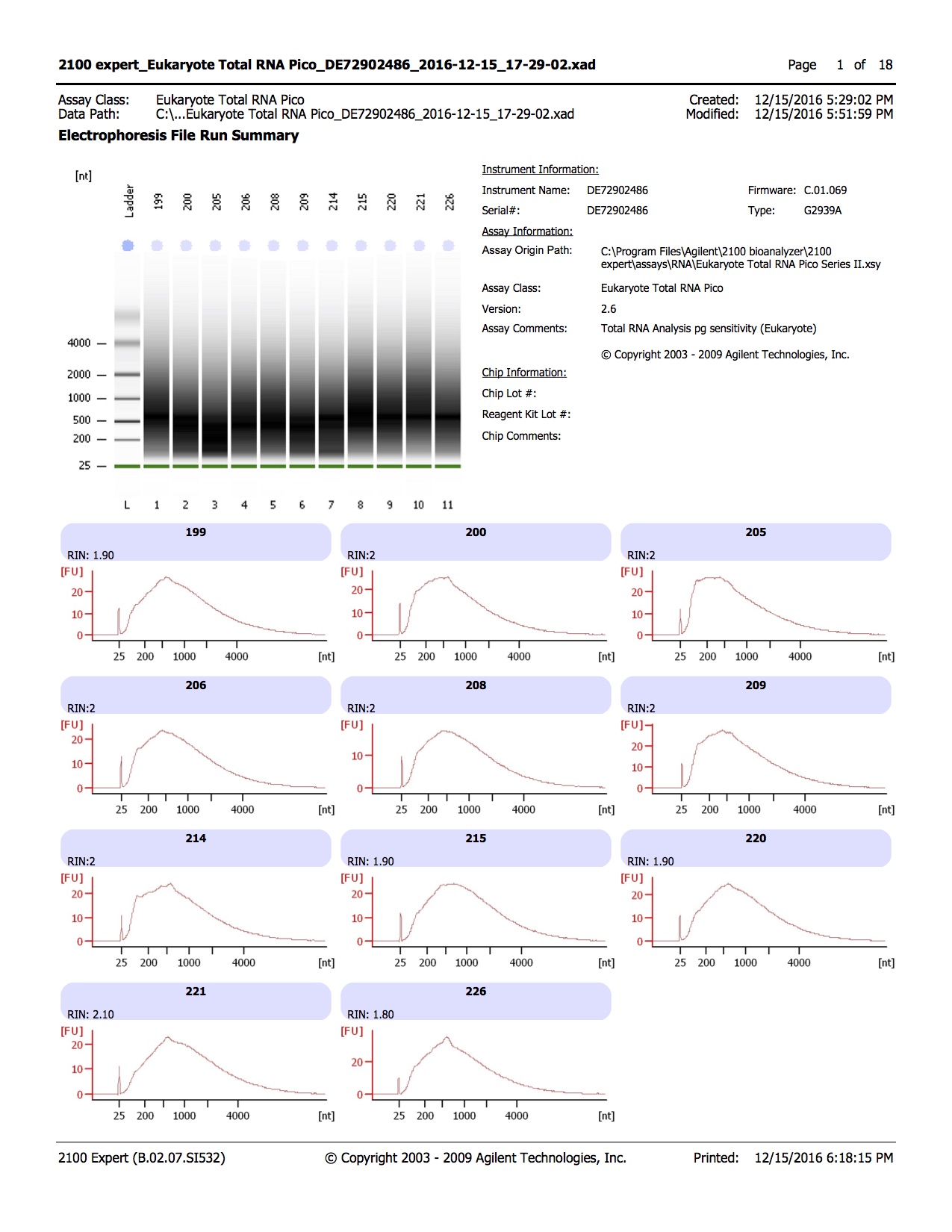
Results of bisulfite converted samples
20161215
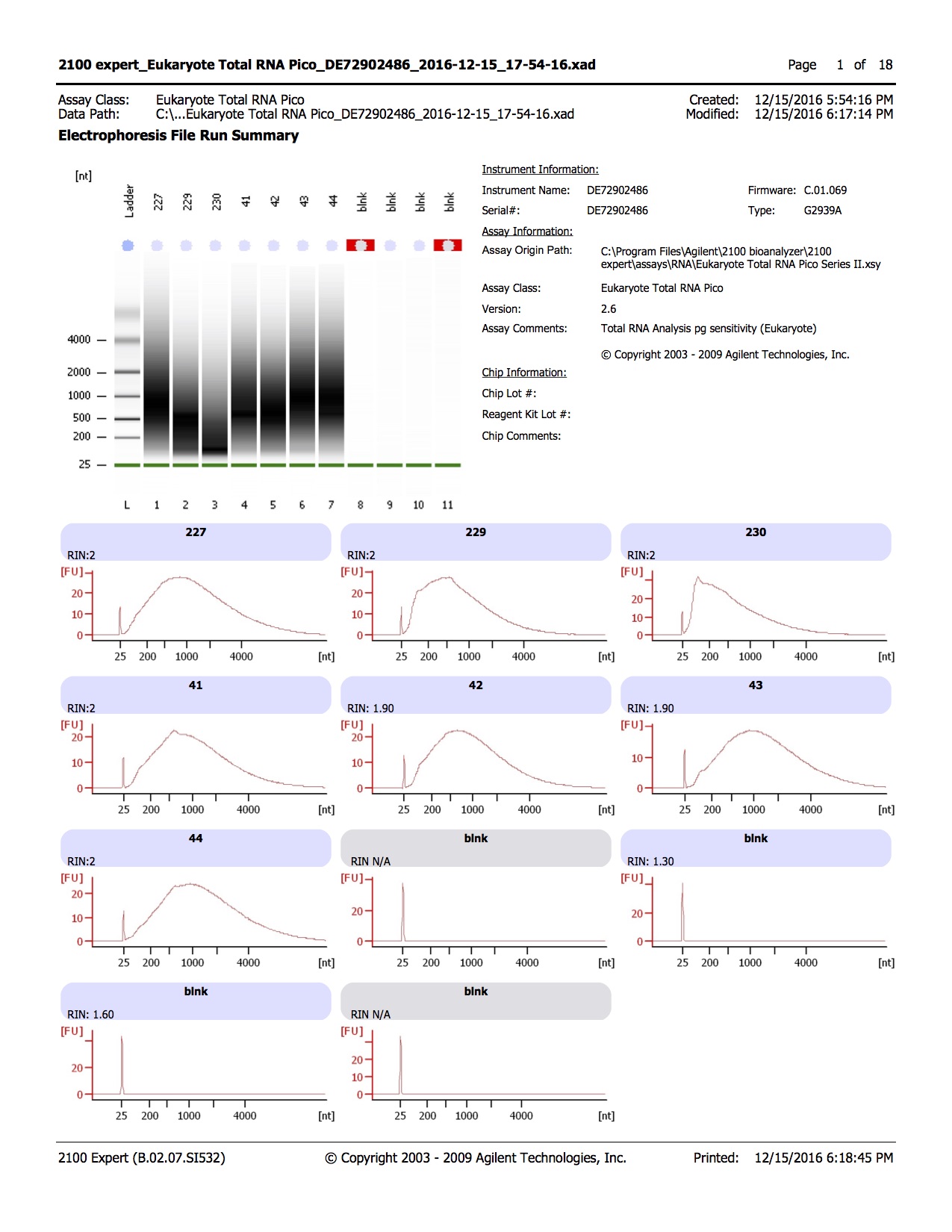
Step 3 Library Prep
20161216
Ilumina TruSeq DNA Methylation Library Preparation Guide
Illumina Adapter Sequences Document # 1000000002694 v01 17 February 2016 TruSeq DNA Methylation Index PCR Primers 5’ CAAGCAGAAGACGGCATACGAGAT[6 bases]GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT
- Index 1: ATCACG
- Index 2: CGATGT
- Index 3: TTAGGC
- Index 4: TGACCA
- Index 5: ACAGTG
- Index 6: GCCAAT
- Index 7: CAGATC
- Index 8: ACTTGA
- Index 9: GATCAG
- Index 10: TAGCTT
- Index 11: GGCTAC
- Index 12: CTTGTA
Illumina library prep was completed according to manufacturer’s instructions with modifications as described in red text on the protocol and listed below.
- samples eluted in 12µl post BS conversion
- 200µl of 80% ethanol used for rinsing beads
- Library prep was started 20161216
- Library prep was finished 20161216
- Individual barcodes were used for each sample from the Illumina TruSeq DNA Methylation Index PCR Primers (10 reactions, 12 indexes Cat #: EGIDX81312)
| Sample.ID | Index.Number | Index.Sequence |
|---|---|---|
| EPI_151 | 1 | ATCACG |
| EPI_152 | 2 | CGATGT |
| EPI_153 | 3 | TTAGGC |
| EPI_154 | 4 | TGACCA |
| EPI_159 | 5 | ACAGTG |
| EPI_160 | 6 | GCCAAT |
| EPI_161 | 7 | CAGATC |
| EPI_162 | 8 | ACTTGA |
| EPI_167 | 9 | GATCAG |
| EPI_168 | 10 | TAGCTT |
| EPI_169 | 11 | GGCTAC |
| EPI_170 | 12 | CTTGTA |
| EPI_175 | 1 | ATCACG |
| EPI_176 | 2 | CGATGT |
| EPI_181 | 3 | TTAGGC |
| EPI_182 | 4 | TGACCA |
| EPI_184 | 5 | ACAGTG |
| EPI_185 | 6 | GCCAAT |
| EPI_187 | 7 | CAGATC |
| EPI_188 | 8 | ACTTGA |
| EPI_193 | 9 | GATCAG |
| EPI_194 | 10 | TAGCTT |
| EPI_199 | 11 | GGCTAC |
| EPI_200 | 12 | CTTGTA |
| EPI_205 | 1 | ATCACG |
| EPI_206 | 2 | CGATGT |
| EPI_208 | 3 | TTAGGC |
| EPI_209 | 4 | TGACCA |
| EPI_214 | 5 | ACAGTG |
| EPI_215 | 6 | GCCAAT |
| EPI_220 | 7 | CAGATC |
| EPI_221 | 8 | ACTTGA |
| EPI_226 | 9 | GATCAG |
| EPI_227 | 10 | TAGCTT |
| EPI_229 | 11 | GGCTAC |
| EPI_230 | 12 | CTTGTA |
| EPI_41 | 1 | ATCACG |
| EPI_42 | 2 | CGATGT |
| EPI_43 | 4 | TGACCA |
| EPI_44 | 8 | ACTTGA |
Library Quantification
20161216 Libraries were quantified on the Qubit using the dsDNA High Sensitivity Kit
- Samples were loaded 1µl of library + 199µl of Qubit dye/buffer mix (1:200)
-
Standards were loaded 10µl of standard + 190µl of Qubit dye/buffer mix (1:20)
- Illumina indicates samples should be >3ng/µl
| Sample.ID | ng/µl |
|---|---|
| EPI_151 | 1.32 |
| EPI_152 | 1.45 |
| EPI_153 | 0.928 |
| EPI_154 | 1.27 |
| EPI_159 | 1.45 |
| EPI_160 | 1.37 |
| EPI_161 | 1.27 |
| EPI_162 | 1.72 |
| EPI_167 | 1.35 |
| EPI_168 | 1.37 |
| EPI_169 | 1.42 |
| EPI_170 | 1.55 |
| EPI_175 | 2.12 |
| EPI_176 | 1.81 |
| EPI_181 | 2.36 |
| EPI_182 | 2.82 |
| EPI_184 | 1.71 |
| EPI_185 | 1.04 |
| EPI_187 | 1.70 |
| EPI_188 | 0.986 |
| EPI_193 | 1.69 |
| EPI_194 | 2.30 |
| EPI_199 | 0.888 |
| EPI_200 | 1.30 |
| EPI_205 | 1.38 |
| EPI_206 | 1.90 |
| EPI_208 | 1.67 |
| EPI_209 | 1.41 |
| EPI_214 | 1.75 |
| EPI_215 | 1.19 |
| EPI_220 | 0.90 |
| EPI_221 | 1.03 |
| EPI_226 | 1.35 |
| EPI_227 | 1.19 |
| EPI_229 | 1.44 |
| EPI_230 | 0.854 |
| EPI_41 | 0.894 |
| EPI_42 | 1.14 |
| EPI_43 | 0.920 |
| EPI_44 | 1.02 |
- Library concentration is lower than expected in the majority of the samples, but higher than previous prep
- llumina states “A total of 3 ng is required to run a sample on an entire HiSeq flow cell (8 lanes).”
Library Quality
20161216
- 1.0µl of each sample was run on the Agilent DNA High Sensitivity Kit
Illumina DNA Methylation kit expected library size on high sensitivity chip
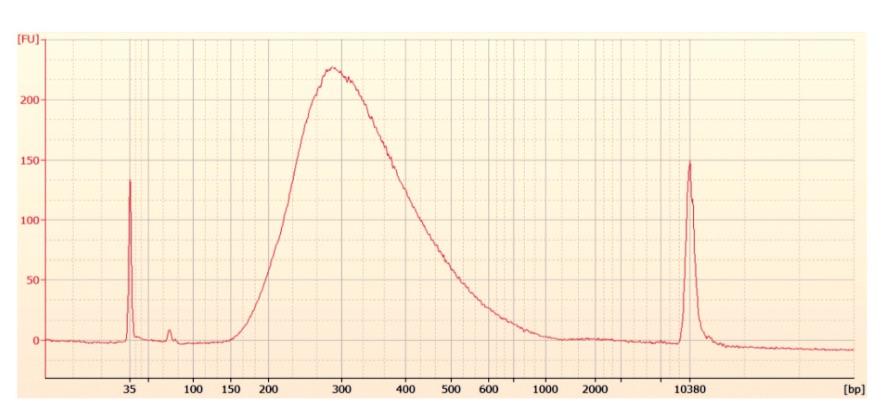
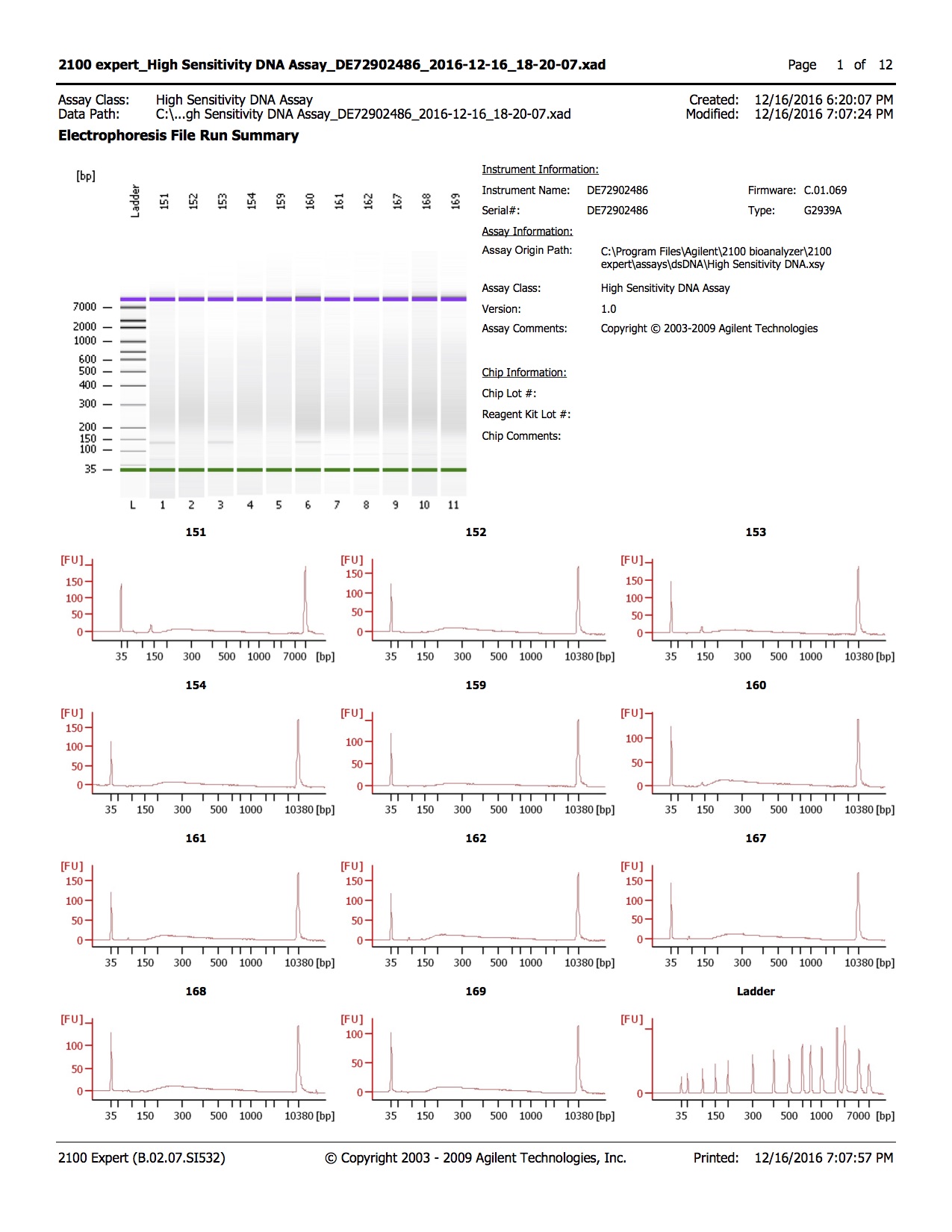
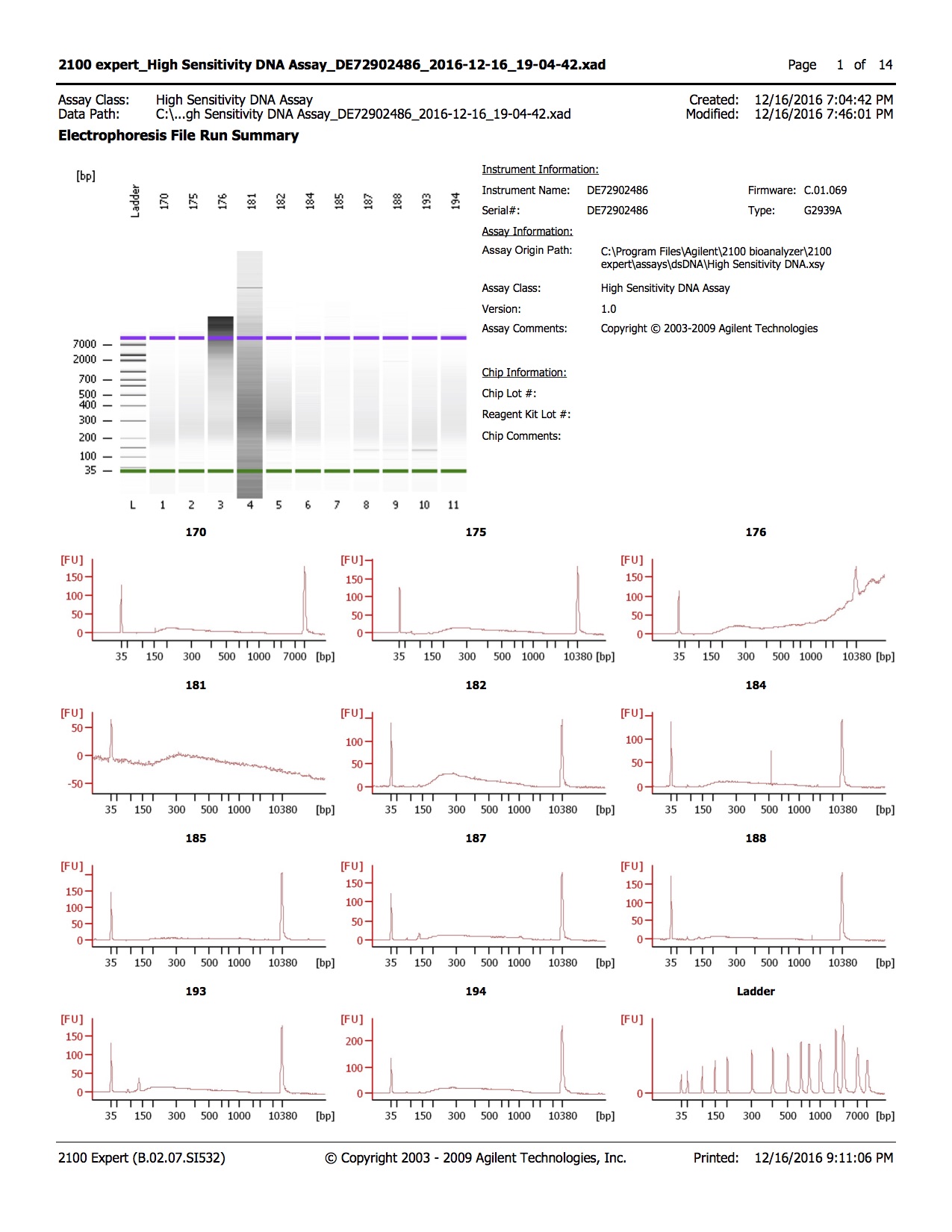
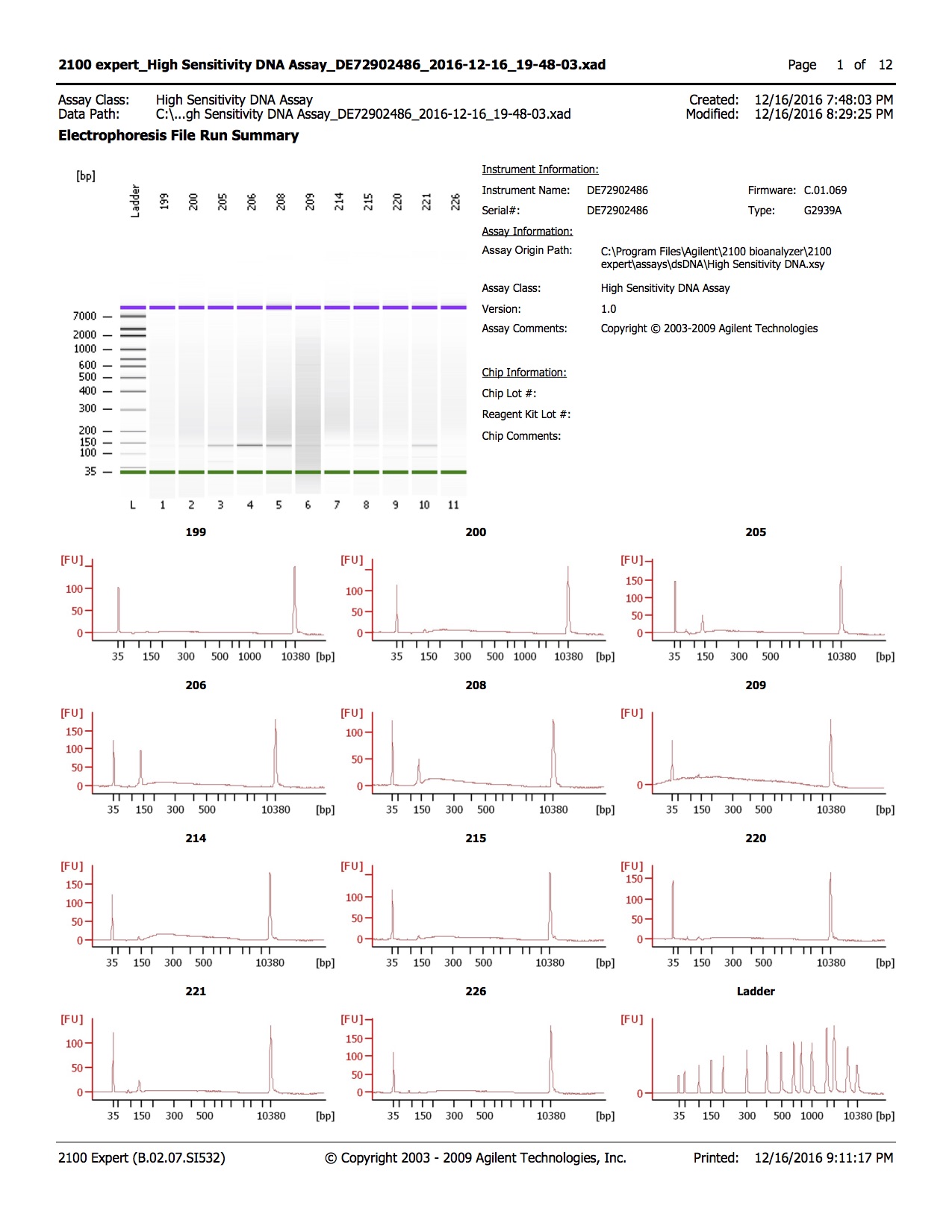
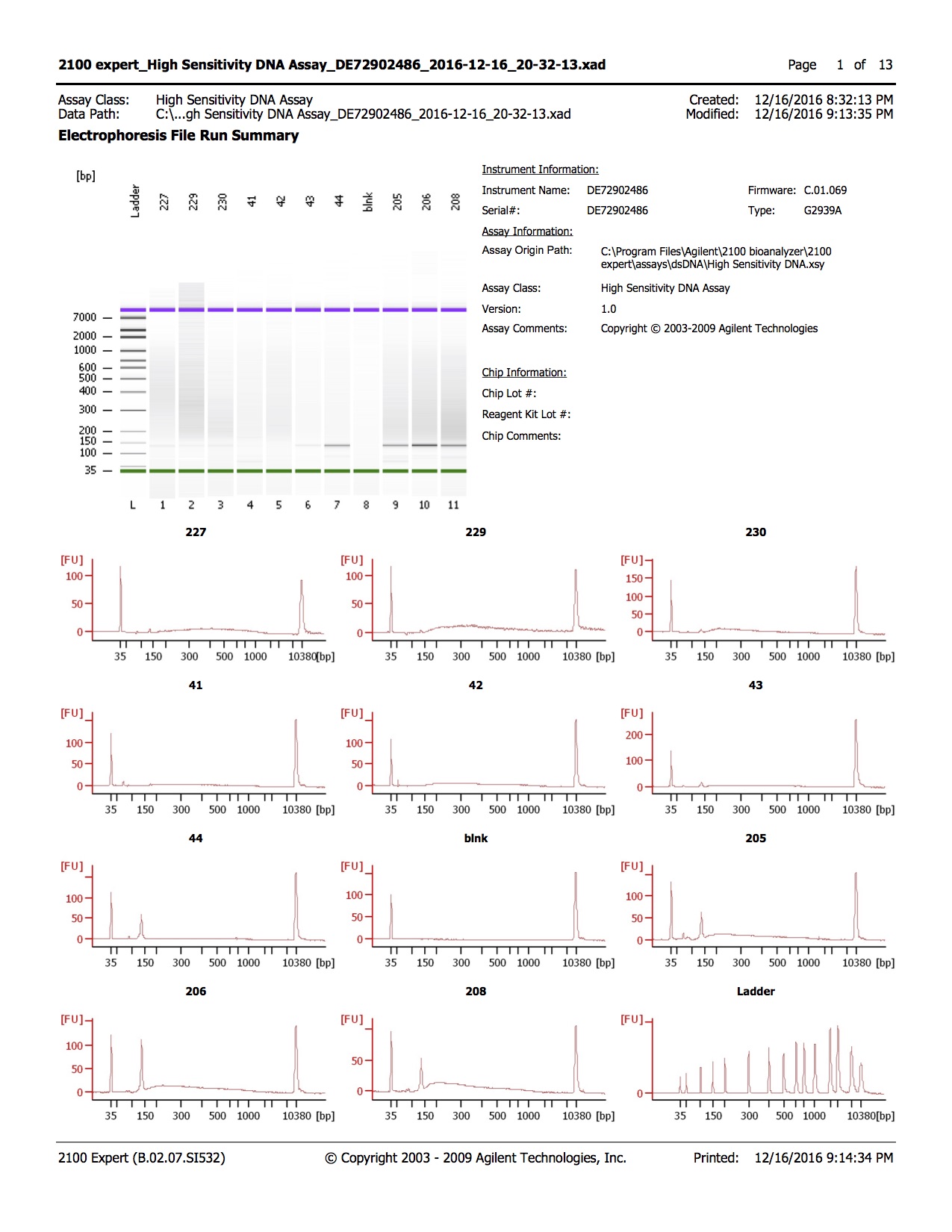
Conclusions
- Library quality appears to be of the right size
- Library quantity is lower than expected, with 1-2ng/µl obtained and ~3ng/µl expected
- Samples 205, 206, and 208 have unexpected peak above lower marker, samples were run twice to confirm.
- Samples are ready for pooling and stored at -20°C
Library Pooling
Samples were pooled by Sam White and sent to Genewiz 20161220 http://onsnetwork.org/kubu4/2016/12/20/sample-submission-geoduck-reduced-representation-bisulfite-pooled-libraries/
Lane 2
| Sample.ID | Index.Number | Index.Sequence |
|---|---|---|
| EPI_151 | 1 | ATCACG |
| EPI_152 | 2 | CGATGT |
| EPI_153 | 3 | TTAGGC |
| EPI_154 | 4 | TGACCA |
| EPI_159 | 5 | ACAGTG |
| EPI_160 | 6 | GCCAAT |
| EPI_161 | 7 | CAGATC |
| EPI_162 | 8 | ACTTGA |
| EPI_167 | 9 | GATCAG |
| EPI_168 | 10 | TAGCTT |
| EPI_169 | 11 | GGCTAC |
| EPI_170 | 12 | CTTGTA |
Lane 3
| Sample.ID | Index.Number | Index.Sequence |
|---|---|---|
| EPI_175 | 1 | ATCACG |
| EPI_176 | 2 | CGATGT |
| EPI_181 | 3 | TTAGGC |
| EPI_182 | 4 | TGACCA |
| EPI_184 | 5 | ACAGTG |
| EPI_185 | 6 | GCCAAT |
| EPI_187 | 7 | CAGATC |
| EPI_188 | 8 | ACTTGA |
| EPI_193 | 9 | GATCAG |
| EPI_194 | 10 | TAGCTT |
| EPI_199 | 11 | GGCTAC |
| EPI_200 | 12 | CTTGTA |
Lane 4
| Sample.ID | Index.Number | Index.Sequence |
|---|---|---|
| EPI_205 | 1 | ATCACG |
| EPI_206 | 2 | CGATGT |
| EPI_208 | 3 | TTAGGC |
| EPI_209 | 4 | TGACCA |
| EPI_214 | 5 | ACAGTG |
| EPI_215 | 6 | GCCAAT |
| EPI_220 | 7 | CAGATC |
| EPI_221 | 8 | ACTTGA |
| EPI_226 | 9 | GATCAG |
| EPI_227 | 10 | TAGCTT |
| EPI_229 | 11 | GGCTAC |
| EPI_230 | 12 | CTTGTA |
Lane 5
| Sample.ID | Index.Number | Index.Sequence |
|---|---|---|
| EPI_41 | 1 | ATCACG |
| EPI_42 | 2 | CGATGT |
| EPI_43 | 4 | TGACCA |
| EPI_44 | 8 | ACTTGA |
| EPI_135 WG | 3 | TTAGGC |
Library QC from Genewiz
Library QC was completed on 20161221 at Genewiz with tapestation and Qubit assays
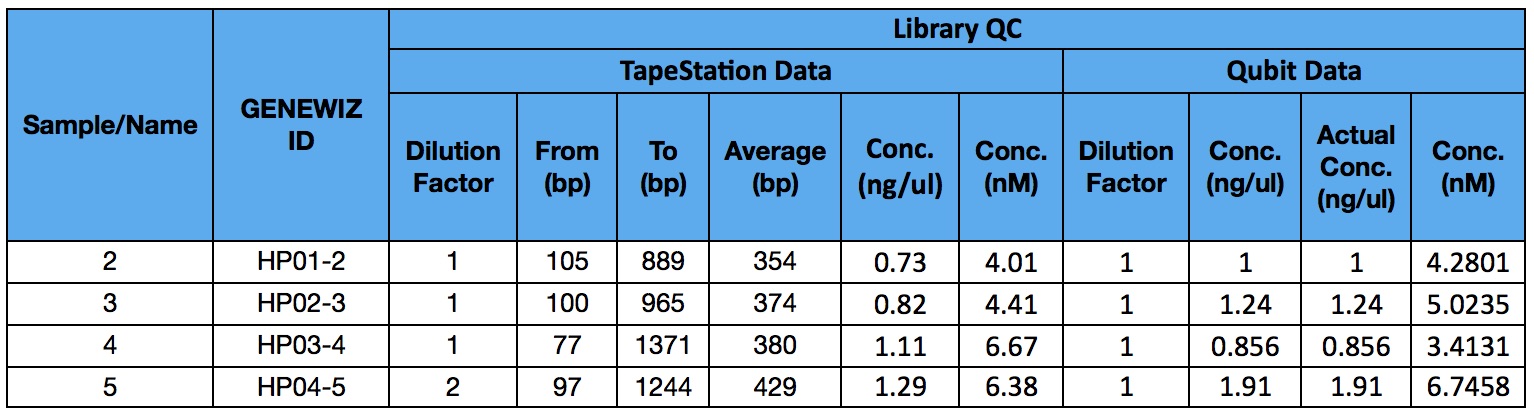
Samples passed QC and were confirmed for sequencing on a single lane of Illumina Highseq2500 2x100bp for each set of 12 and for the last set of 5 on 20161222.
