Chelex DNA Extraction
Testing Chelex DNA extraction for rapid DNA acquisition for Pocillopora ID in the field
Pocillopora ID Johnston et al 2018 RFLP
Extraction adapted from SAKH protocol from Simon et al. (2020) Biology Methods and Protocols, 5, bpaa009
l
Chelex 100 molecular biology grade resin BioRad Cat #1421253
Pipettes: 20-200uL single or multichannel, based on your preference
Pipette tips: Enough to transfer 100uL of chelex into strip tubes or plate
Bone or heavy-duty nail cutters to clip coral samples
Forceps to transfer sample into chelex solution
1.5ml microfuge tubes
Isopropanol wipes and RO or DI water (for rinsing cutters and forceps between samples)
Thermal cycler or heat block
Lab vortex mixer
Centrifuge for 1.5ml tubes that can reach 13,000-14,000 rpm
Agarose
1X TAE
Gel dye
Gel ladder GeneRuler 100 bp DNA Ladder (Thermo FIsher Catalog SM0241)
Gel ladder GeneRuler 1kb plus DNA Ladder (Thermo FIsher Catalog SM1333)
DNA Samples
Extraction Protocol Steps
Chelex Preparation
Prepare 5% Chelex
- Make in small batches of ~45 mL (2.25 g chelex in 45mL of MilliQ H2O)
Pre-heat 5% Chelex solution to 95˚C prior to use for extraction
Microcentrifuge tube (1.5mL) Chelex DNA extraction protocol
- Make sure Chelex is well-mixed and each tube receives chelex resin (the pipet tip should look a little cloudy with Chelex solution) - Always include a negative control tube with no sample
- clipped 4 small 4mm^2 samples from Pocillopora acuta from the CBLS wetlab and added to 4/6 1.5ml microfuge tubes
- Sample ID (H= Hollie Putnam, P = Pierrick Harnay)
- H1=Pacuta clip1
- H2=Pacuta clip2
- H3= Negative control
- P1=Pacuta clip3
- P2=Pacuta clip4
- P3=Negative control
- Sample ID (H= Hollie Putnam, P = Pierrick Harnay)
- Added 200 µL of pre-heated (95˚C) 5% Chelex
- Vortex for 30 sec
a. Made sure tissue is still in the Chelex solution after vortexing - Incubated at 95˚C for 15 minutes, vortexing every 5 min and also used the Eppendorf heated mixer at 500prm
- Centrifuged for 3 min at 14,000 rpm to pellet Chelex and tissue (benchtop centrifuge)
- Pipetted ~180 µL of supernatant to a new 1.5 mL tube
- Centrifuged for 3 min at 14,000 rpm to pellet Chelex and tissue
- Pipeted 150 µL of supernatant to a new, labeled 1.5 mL tube
a. This is the final tube of extracted DNA so we labelled with sample name on the lid and the sample name, extraction date, and initials on the side
b. We stored at 4˚C for immediate use with the following gel, and the remainder was stored at -20˚C.
Gel Electrophoresis of the DNA on the Bento Lab
Bento Lab Gel Casting Instructions
- Prepared a 1% agarose gel in 1x TAE with a 9 well comb from the Bento Lab
a. Melted 0.5g of agarose in 50ml of 1X TAE and poured gel - Used 0.5X TAE as running buffer
- Mixed 1.5µl of 6x tracking/loading dye with 4µl of each sample except 1kb ladder which was already dyed.
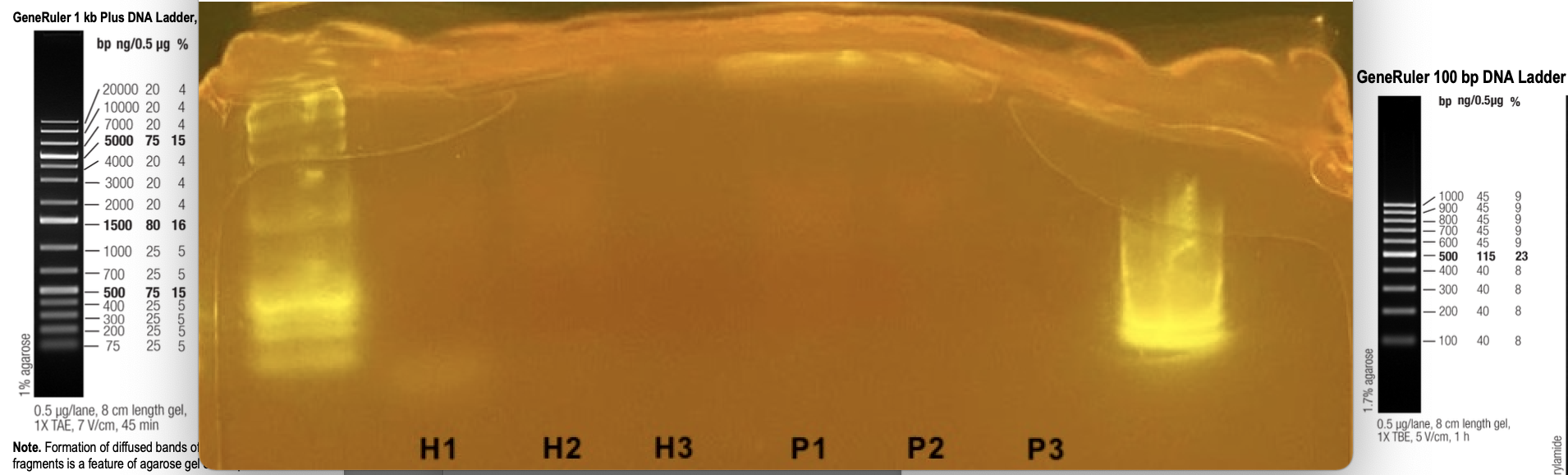
- Added two ladders and 6 samples in the following order:
Well 1. 10kb GeneRuler ladder
Well 2.H1
Well 3.H2
Well 4.H3
Well 5.P1
Well 6.P2
Well 7.P3
Well 8.100bp GeneRuler ladder - Ran the gel for 45 minutes at 50V
Gel Results
-
TAE gels are consistently melting at the top in the Bento Lab eletrophoresis chamber
-
Ladders are difficult to discern
-
DNA appears to be present in a smear around 4000-1000bp
-
H1 also appears to have a small band <75bp
-
H3 negative control appears to have no DNA
Next steps
Use the extracted DNA and compare amplification of the mtORF region using the extracted DNA and direct samples with both:
FERK1071 DREAMTAQ Master Mix
F170S PHIRE TISSUE DIRECT PCR Master Mix
