Direct mtORF PCR Test
Testing Chelex-extracted DNA in comparison to direct PCR of tissue for Pocillopora ID in the field
Equipment and Reagents
- Bento Lab Bento Lab thermal cycler, gel, centrifuge
- BSA Thermo Scientific™ Pierce™ Bovine Serum Albumin Standard Ampules, 2mg/ml, Catalog #PI23209
- F primer FatP6.1 200µM Stock IDT)
- R primer RORF 200µM Stock IDT
- Gel ladder GeneRuler 1kb plus DNA Ladder (Thermo FIsher Catalog SM1333)
- Master Mix FERK1071 DREAMTAQ Master Mix
- Direct Master Mix F170S PHIRE TISSUE DIRECT PCR Master Mix
- Gel Stain Biotium GelGreen Nucleic Acid Gel Stain, 10,000X in Water Fisher Cat NC9728313
Prepare Reagents
- diluted BSA from 2mg/ml to 0.25mg/ml in Nuclease Free water
- F primer FastP6.1 (Added 50µl of stock 2mg/ml plus 950µl of Nuclease Free water and made 3 aliquots of ~320µl each and stored in reagent box at -20)
- R primer RORF (Added 50µl of stock 2mg/ml plus 950µl of Nuclease Free water and made 3 aliquots of ~320µl each and stored in reagent box at -20)
Chelex DNA Extraction was completed on April 9th 2022 and DNA stored at -20°C
TESTING mtORF AMPLIFICATION OF CHELEX EXTRACTED DNA FROM 4/9/22
Dream Taq PCR Master Mix (2x)
Master Mix:
| Reagent | 1Rxn µl | 10 Rxn µl |
|---|---|---|
| Phire Taq Mix | 7.5 | 75 |
| BSA (0.25mg/ml) | 0.14 | 1.4 |
| F primer FastP6.1 (10µM) | 0.195 | 1.95 |
| R primer RORF | 0.195 | 1.95 |
| DNA | 1.5 | NA |
| H2O | 5.47 | 54.7 |
| Total volume | 15 | 150 |
Thermal Cycling Conditions
- [95°C 60 secondes] 1 cycle
- [95°C 30 sec,53°C 30 sec, 72°C 70 sec] 40 cycles
- [72°C 5 minutes] 1 cycle
- [15°C infinity]
Sample
1: H1 Hollie 1 chelex extraction of P. acuta 4/9/22
2: H2 Hollie 2 chelex extraction of P. acuta 4/9/22
3: H3 Hollie negative extraction control 4/9/22
4: P1 Pierrick 1 chelex extraction of P. acuta 4/9/22
5: P2 Pierrick 2 chelex extraction of P. acuta 4/9/22
6: P3 Pierrick negative extraction control 4/9/22
7: H20 Negative PCR Control
8: 253= Pocillopora gDNA from E5 Terpis
Gel protocol
prepared a 50ml gel using 50ml of 1xTAE + 0.5g Agarose, melted gel for ~90 sec in the microwave and let cool and added 1µl Gelgreen
Ran the gel in 0.25X TAE to reduce heating in the Bento Lab, but had to move the gel to on of the OWl rigs in order to start the next PCR in the Bento Lab.
Gel Results
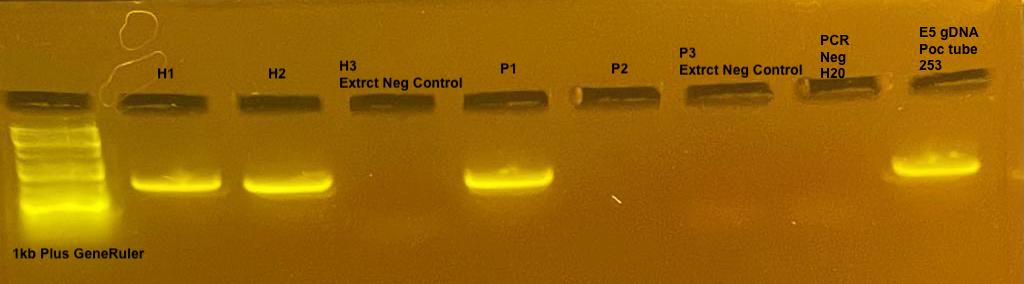
3 of the 4 coral samples extracted with Chelex amplified the desired band with the mtORF primers along with the positive control
TESTING mtORF AMPLIFICATION DIRECT FROM CORAL SAMPLES
Phire Taq Mix Direct PCR
Phire Master Mix:
| Reagent | 1Rxn µl | 7 Rxn µl |
|---|---|---|
| Phire Taq Mix | 7.5 | 52.5 |
| BSA (0.25mg/ml) | NA | NA |
| F primer FastP6.1 (10µM) | 0.195 | 1.365 |
| R primer RORF (10µM) | 0.195 | 1.365 |
| DNA | Direct Clipping | Direct Clipping |
| H2O | 7.11 | 49.77 |
| Total volume | 15 | 105 |
Aliquot 13.5µl of master mix into each tube
Phire Thermal Cycling Conditions
- [98°C 5 minutes] 1 cycle
- [98°C 10 sec, 58°C 10 sec, 72°C 20 sec] 40 cycles
- 72°C 1 minute 1 cycle
- 15°C infinity
- The protocol calls for 5 second steps, but the Bento Lab shortest step time is 10seconds
Phire Samples
scraped samples from the coral plug P. acuta before scraping all 3 samples (left) and after (right)
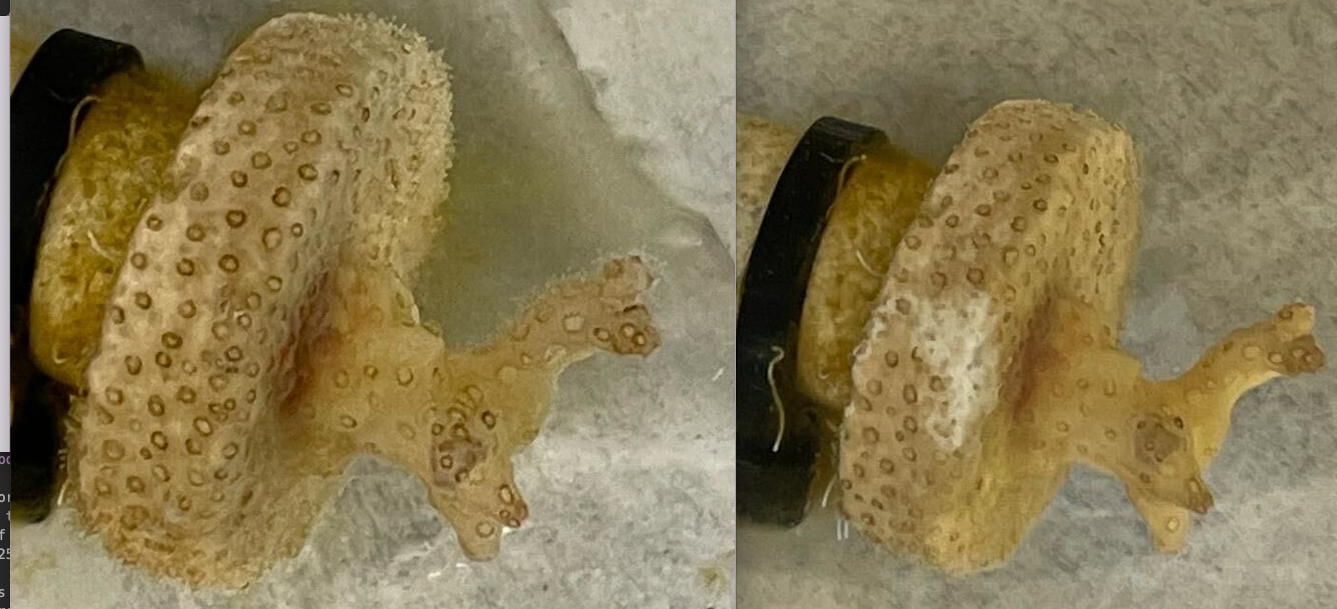
1: Cl= Clip1 - largest piece ~2mm x 2mm
2: C2= Clip2 - piece 1mm x 1mm
3: C3= Clip3 - piece 0.5mm x 0.5mm
4: Neg= H2O
5: H1= Chelex 1
6: 253= gDNA 253 Poc from E5 Terpis
7: C4= Clip4 - Scraped w/pipette tip only
8: P2 from above run again
Phire Gel protocol
prepared a 50ml gel using 50ml of 1xTAE + 0.5g Agarose, melted gel for ~90 sec in the microwave and let cool and added 1µl Gelgreen
Ran the gel in 0.25X TAE to reduce heating in the Bento Lab, but gel melting was still visible.
Phire Gel Results
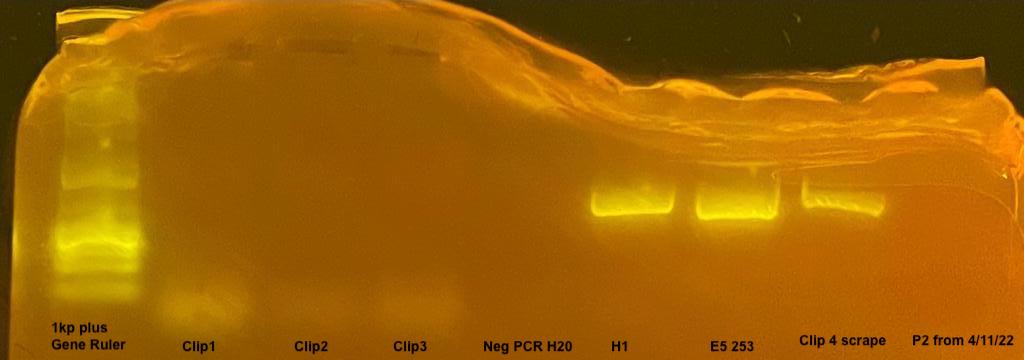
The clippings for the coral inhibited the PCR, the only sample that worked for the direct from tissue PCR was the “Clip 4” which was only coral tissue rubbed on the side of the plug with the pipette tip. The chelex sample H1 and the gDNA E5 sample 253 also worked.
It seems less is more in this case, so this bodes well for taking swabs of the coral DNA from recruitment plates.
