Poc ID via mtORF POC RAPID Plate 002 Plate 003
gDNA extractions for Pocillopora species ID COTS POC RAPID Plate002 and Plate003
Equipment and Reagents
- Sample Preservation and Lysis Zymo Research DNA/RNA Shield R1100-250
- DNA Extraction 96 Well Plate Zymo Research Quick-DNA™ 96 D3010
- DNA Extraction Individual Samples Zymo Research Quick-DNA™ Miniprep plus kit extraction Cat D4068
- Proteinase K Zymo Proteinase K with Storage BufferZymo D3001-2-20
- PK Digestion Buffer Zymo PK Digestion Buffer R1200-1-20
- Foward primer FatP6.1 200µM Stock IDT
- Reverse primer RORF 200µM Stock IDT
- Master Mix EmeraldAmp GT PCR Master Mix RR320A
- EmeraldAmp GT PCR Master Mix is a loading-dye-added version of EmeraldAmp MAX PCR Master Mix that is optimized for great performance and convenience in both standard and high-throughput PCR applications.
- Loading Dye NEB 6X Purple Loading Dye NEB Cat # B7024S
- Gel Stain Biotium GelGreen Nucleic Acid Gel Stain, 10,000X in Water Fisher Cat NC9728313
- DNA Ladder Image NEB Cat N0550G 1kb Gel ladder NEB Cat N0550G
- DNA Ladder NEB Cat N0550G 1kb Gel ladder NEB Cat N0550G
- KapaPure Beads Fisher 50-196-5220 Roche Diagnostics 07983280001
- Eppendorf Deepwell Plate 96/2000 µL plates Eppendorf Cat # 951033502
23 April 2024
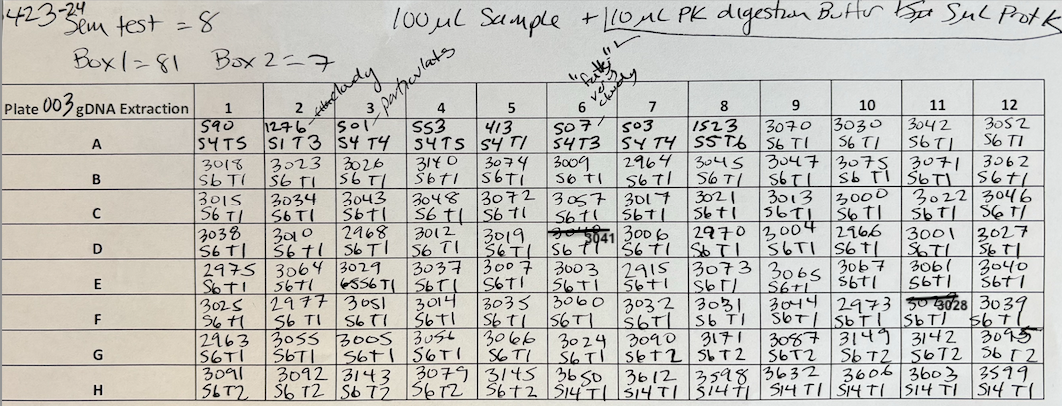
Plate preparation Plate 003
Moved 100µl of sample liquid (lysed sample in Zymo RNA/DNA Shield) to deep well 96 eppendorf plate. Added 10µl of PK DIgestion buffer and 5µl of ProteinaseK to each well and mixed by pipetting. Sealed plate with a foil cover and placed at -20°C for subsequent extraction.
Samples
24 April 2024
DNA Extraction
DNA was extracted withZymo Research Quick-DNA™ 96 Kit D3010
Used Page 10 Proteinase K Digestion with DNA/RNA Shield
Used Eppendorf Deepwell Plate 96/2000 µL plates Catalog No. 951033502 for reagent and sample mixing prior to adding to Zymo column plate
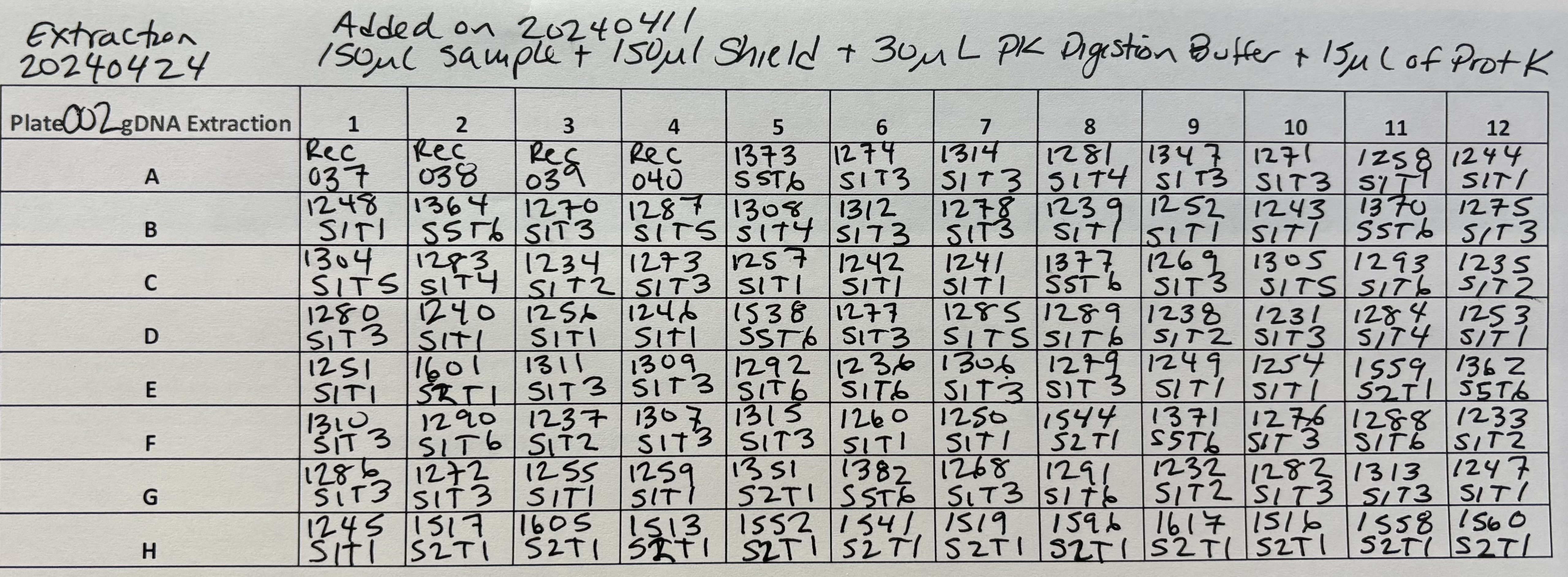
Plate 002 was already prepared in the deep well plate from the11April2024 Prep
The samples were extracted according to the manufacturer’s instructions for the Quick DNA 96 Kit for samples stored in DNA/RNA Shield including the addition of Proteinase K (20mg ml-1).
For Plate 002
- 4 volumes of Zymo Kit Genomic Lysis Buffer was added for each volume of the sample digestion (e.g., add 1200µl of Genomic Lysis Buffer to 300µl of sample digestion). Samples were mixed via pipetting and the mixtures were incubated for 5 minutes at room temperature.
Each well of the Silicon-A plate set on the collection plate can hold 600µl, but this brings the liquid too close to the top and increases chances of carry over to other wells. Becasue of this, I added the sample in 3 steps for Plate 002:
1) 400µl and then 5 minute spin at 2500 rcf
2) 400µl and then 5 minute spin at 2500 rcf
3) 400µl and then 5 minute spin at 2500 rcf
For Plate 003 Added 100µl of sample + 10µl of PK Digestion Buffer + 5µl of ProteinaseK. Pipetted to mix and incubated for 5 min at room temp.
- 4 volumes of Zymo Kit Genomic Lysis Buffer was added for each volume of the sample digestion (e.g., add 400µl of Genomic Lysis Buffer to 100µl of sample digestion). Samples were mixed via pipetting and the mixtures were incubated for 5 minutes at room temperature. Then samples were moved to the plate columns. I added 400µl and then 5 minute spin at 2500 rcf.
The waste was removed from the collection plate after each centrifugation step.
Once all steps were complete to this point for both Plate 002 and Plate 003, all steps were completed the same for both plates as described below.
-
Next 200µl of DNA Pre-wash buffer was added to each well and then the plate was spun for 5 minute at 2500 rcf
-
Next 300µl of g-DNA wash buffer was added to each well and then the plate was spun for 5 minute at 2500 rcf
-
DNA was then eluted in 50µl of kit Elution Buffer warmed to 70°C. gDNA was stored at -20°C in the elution plate covered with a foil seal.
24 April 2024
Quantification of gDNA
Used Lane lab nanodrop and kit elution buffer as blank to quantify the first column of DNA on the plate. Used 1.5µl of sample
| Sample ID | Well | Project | ng/µl | A260/280 | A260/230 | gDNA Plate |
|---|---|---|---|---|---|---|
| 590 S4 T5 | A1 | POC COTS RAPID RA | 17.9 | 2.07 | 1.90 | Plate003 |
| 3018 S6 T1 | B1 | POC COTS RAPID RA | 18.8 | 2.00 | 1.85 | Plate003 |
| 3015 S6 T1 | C1 | POC COTS RAPID RA | 7.8 | 2.31 | 0.81 | Plate003 |
| Rec-037 | A1 | TPC COTS RAPID | 5.8 | 2.35 | 2.71 | Plate002 |
| 1248 S1T1 | B1 | POC COTS RAPID RA | 6.8 | 2.65 | 1.78 | Plate002 |
| 1304 S1T5 | C1 | POC COTS RAPID RA | 33.3 | 1.95 | 1.44 | Plate002 |
All tested samples had DNA and can move forward for PCR.
